Machine-learning algorithms define pathogen-specific local immune fingerprints in peritoneal dialysis patients with bacterial infections
- PMID: 28318629
- PMCID: PMC5484022
- DOI: 10.1016/j.kint.2017.01.017
Machine-learning algorithms define pathogen-specific local immune fingerprints in peritoneal dialysis patients with bacterial infections
Abstract
The immune system has evolved to sense invading pathogens, control infection, and restore tissue integrity. Despite symptomatic variability in patients, unequivocal evidence that an individual's immune system distinguishes between different organisms and mounts an appropriate response is lacking. We here used a systematic approach to characterize responses to microbiologically well-defined infection in a total of 83 peritoneal dialysis patients on the day of presentation with acute peritonitis. A broad range of cellular and soluble parameters was determined in peritoneal effluents, covering the majority of local immune cells, inflammatory and regulatory cytokines and chemokines as well as tissue damage-related factors. Our analyses, utilizing machine-learning algorithms, demonstrate that different groups of bacteria induce qualitatively distinct local immune fingerprints, with specific biomarker signatures associated with Gram-negative and Gram-positive organisms, and with culture-negative episodes of unclear etiology. Even more, within the Gram-positive group, unique immune biomarker combinations identified streptococcal and non-streptococcal species including coagulase-negative Staphylococcus spp. These findings have diagnostic and prognostic implications by informing patient management and treatment choice at the point of care. Thus, our data establish the power of non-linear mathematical models to analyze complex biomedical datasets and highlight key pathways involved in pathogen-specific immune responses.
Keywords: biomarkers; inflammation; machine learning methods; microbial infection; peritoneal dialysis.
Copyright © 2017 International Society of Nephrology. Published by Elsevier Inc. All rights reserved.
Figures

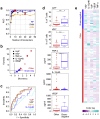

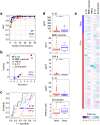

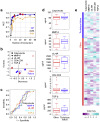
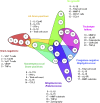
Comment in
-
Understanding the immune signature fingerprint of peritoneal dialysis-related peritonitis.Kidney Int. 2017 Jul;92(1):16-18. doi: 10.1016/j.kint.2017.02.027. Kidney Int. 2017. PMID: 28646991
References
-
- Janeway C.A., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;541:1–13. - PubMed
-
- Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. - PubMed
-
- Blander J.M., Sander L.E. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. 2012;12:215–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

