Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii
- PMID: 28319081
- PMCID: PMC7616673
- DOI: 10.1038/nmicrobiol.2017.34
Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii
Abstract
Prokaryotic Argonaute proteins acquire guide strands derived from invading or mobile genetic elements, via an unknown pathway, to direct guide-dependent cleavage of foreign DNA. Here, we report that Argonaute from the archaeal organism Methanocaldococcus jannaschii (MjAgo) possesses two modes of action: the canonical guide-dependent endonuclease activity and a non-guided DNA endonuclease activity. The latter allows MjAgo to process long double-stranded DNAs, including circular plasmid DNAs and genomic DNAs. Degradation of substrates in a guide-independent fashion primes MjAgo for subsequent rounds of DNA cleavage. Chromatinized genomic DNA is resistant to MjAgo degradation, and recombinant histones protect DNA from cleavage in vitro. Mutational analysis shows that key residues important for guide-dependent target processing are also involved in guide-independent MjAgo function. This is the first characterization of guide-independent cleavage activity for an Argonaute protein potentially serving as a guide biogenesis pathway in a prokaryotic system.
Figures

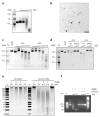
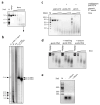
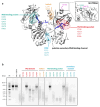
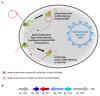
Comment in
-
Archaeal physiology: Two modes of a DNA scissor.Nat Microbiol. 2017 May 25;2:17049. doi: 10.1038/nmicrobiol.2017.49. Nat Microbiol. 2017. PMID: 28540923 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

