Human haematopoietic stem cell lineage commitment is a continuous process
- PMID: 28319093
- PMCID: PMC5496982
- DOI: 10.1038/ncb3493
Human haematopoietic stem cell lineage commitment is a continuous process
Abstract
Blood formation is believed to occur through stepwise progression of haematopoietic stem cells (HSCs) following a tree-like hierarchy of oligo-, bi- and unipotent progenitors. However, this model is based on the analysis of predefined flow-sorted cell populations. Here we integrated flow cytometric, transcriptomic and functional data at single-cell resolution to quantitatively map early differentiation of human HSCs towards lineage commitment. During homeostasis, individual HSCs gradually acquire lineage biases along multiple directions without passing through discrete hierarchically organized progenitor populations. Instead, unilineage-restricted cells emerge directly from a 'continuum of low-primed undifferentiated haematopoietic stem and progenitor cells' (CLOUD-HSPCs). Distinct gene expression modules operate in a combinatorial manner to control stemness, early lineage priming and the subsequent progression into all major branches of haematopoiesis. These data reveal a continuous landscape of human steady-state haematopoiesis downstream of HSCs and provide a basis for the understanding of haematopoietic malignancies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

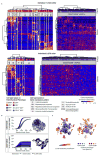
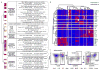
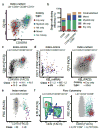




Comment in
-
Demystifying blood stem cell fates.Nat Cell Biol. 2017 Mar 31;19(4):261-263. doi: 10.1038/ncb3494. Nat Cell Biol. 2017. PMID: 28361939
-
Hematopoiesis Lineage Tree Uprooted: Every Cell Is a Rainbow.Dev Cell. 2017 Apr 10;41(1):7-9. doi: 10.1016/j.devcel.2017.03.020. Dev Cell. 2017. PMID: 28399402 Free PMC article.
References
-
- Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. - PubMed
-
- Morrison S, Uchida N, Weissman I. The biology of hematopoietic stem cells. Annu Rev Cell Dev Biol. 1995;11:35–71. - PubMed
-
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. - PubMed
-
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. - PubMed
-
- Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

