Accurate identification of single-nucleotide variants in whole-genome-amplified single cells
- PMID: 28319112
- PMCID: PMC5408311
- DOI: 10.1038/nmeth.4227
Accurate identification of single-nucleotide variants in whole-genome-amplified single cells
Abstract
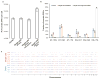
Mutation analysis in single-cell genomes is prone to artifacts associated with cell lysis and whole-genome amplification. Here we addressed these issues by developing single-cell multiple displacement amplification (SCMDA) and a general-purpose single-cell-variant caller, SCcaller (https://github.com/biosinodx/SCcaller/). By comparing SCMDA-amplified single cells with unamplified clones from the same population, we validated the procedure as a firm foundation for standardized somatic-mutation analysis in single-cell genomics.
Conflict of interest statement
X.D., L.Z., M.L., A.M. and J.V. are co-founders of SingulOmics Corp.
Figures
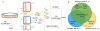

References
-
- Fryxell KJ, Zuckerkandl E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Molecular biology and evolution. 2000;17:1371–1383. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

