High-throughput transformation of Saccharomyces cerevisiae using liquid handling robots
- PMID: 28319150
- PMCID: PMC5358765
- DOI: 10.1371/journal.pone.0174128
High-throughput transformation of Saccharomyces cerevisiae using liquid handling robots
Abstract
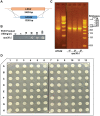
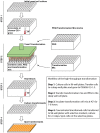
Saccharomyces cerevisiae (budding yeast) is a powerful eukaryotic model organism ideally suited to high-throughput genetic analyses, which time and again has yielded insights that further our understanding of cell biology processes conserved in humans. Lithium Acetate (LiAc) transformation of yeast with DNA for the purposes of exogenous protein expression (e.g., plasmids) or genome mutation (e.g., gene mutation, deletion, epitope tagging) is a useful and long established method. However, a reliable and optimized high throughput transformation protocol that runs almost no risk of human error has not been described in the literature. Here, we describe such a method that is broadly transferable to most liquid handling high-throughput robotic platforms, which are now commonplace in academic and industry settings. Using our optimized method, we are able to comfortably transform approximately 1200 individual strains per day, allowing complete transformation of typical genomic yeast libraries within 6 days. In addition, use of our protocol for gene knockout purposes also provides a potentially quicker, easier and more cost-effective approach to generating collections of double mutants than the popular and elegant synthetic genetic array methodology. In summary, our methodology will be of significant use to anyone interested in high throughput molecular and/or genetic analysis of yeast.
Conflict of interest statement
Figures


Similar articles
-
Rapid Prototyping Platform for Saccharomyces cerevisiae Using Computer-Aided Genetic Design Enabled by Parallel Software and Workcell Platform Development.SLAS Technol. 2019 Jun;24(3):291-297. doi: 10.1177/2472630318798304. Epub 2018 Aug 30. SLAS Technol. 2019. PMID: 30165777
-
Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method.Nat Protoc. 2007;2(1):38-41. doi: 10.1038/nprot.2007.15. Nat Protoc. 2007. PMID: 17401336
-
High-throughput yeast plasmid overexpression screen.J Vis Exp. 2011 Jul 27;(53):2836. doi: 10.3791/2836. J Vis Exp. 2011. PMID: 21841759 Free PMC article.
-
Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries.Methods Mol Biol. 2011;781:127-59. doi: 10.1007/978-1-61779-276-2_8. Methods Mol Biol. 2011. PMID: 21877281 Review.
-
Robotic liquid handling and automation in epigenetics.J Lab Autom. 2012 Oct;17(5):327-9. doi: 10.1177/2211068212457160. Epub 2012 Aug 29. J Lab Autom. 2012. PMID: 22933618 Review.
Cited by
-
CRISPR-UnLOCK: Multipurpose Cas9-Based Strategies for Conversion of Yeast Libraries and Strains.Front Microbiol. 2017 Sep 20;8:1773. doi: 10.3389/fmicb.2017.01773. eCollection 2017. Front Microbiol. 2017. PMID: 28979241 Free PMC article.
-
Transcription factors induce differential splicing of duplicated ribosomal protein genes during meiosis.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1321. doi: 10.1093/nar/gkae1321. Nucleic Acids Res. 2025. PMID: 39817510 Free PMC article.
-
Accelerated Bioprocess Development of Endopolygalacturonase-Production with Saccharomyces cerevisiae Using Multivariate Prediction in a 48 Mini-Bioreactor Automated Platform.Bioengineering (Basel). 2018 Nov 21;5(4):101. doi: 10.3390/bioengineering5040101. Bioengineering (Basel). 2018. PMID: 30469407 Free PMC article.
-
CATS: Cas9-assisted tag switching. A high-throughput method for exchanging genomic peptide tags in yeast.BMC Genomics. 2020 Mar 10;21(1):221. doi: 10.1186/s12864-020-6634-9. BMC Genomics. 2020. PMID: 32156257 Free PMC article.
-
Implications of Microorganisms in Alzheimer's Disease.Curr Issues Mol Biol. 2022 Sep 30;44(10):4584-4615. doi: 10.3390/cimb44100314. Curr Issues Mol Biol. 2022. PMID: 36286029 Free PMC article. Review.
References
-
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285: 901–906. - PubMed
-
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, et al. Functional Discovery via a Compendium of Expression Profiles. Cell. 2000;102: 109–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

