EnCOUNTer: a parsing tool to uncover the mature N-terminus of organelle-targeted proteins in complex samples
- PMID: 28320318
- PMCID: PMC5359831
- DOI: 10.1186/s12859-017-1595-y
EnCOUNTer: a parsing tool to uncover the mature N-terminus of organelle-targeted proteins in complex samples
Abstract
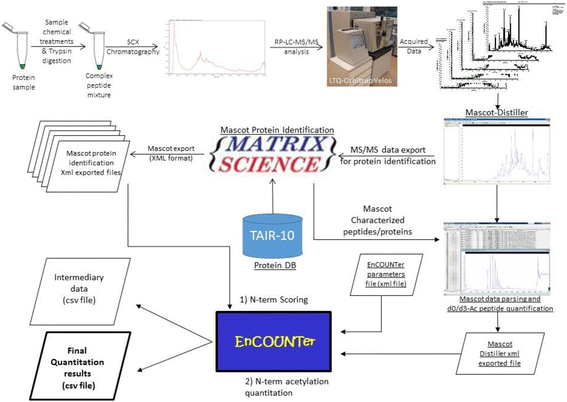
Background: Characterization of mature protein N-termini by large scale proteomics is challenging. This is especially true for proteins undergoing cleavage of transit peptides when they are targeted to specific organelles, such as mitochondria or chloroplast. Protein neo-N-termini can be located up to 100-150 amino acids downstream from the initiator methionine and are not easily predictable. Although some bioinformatics tools are available, they usually require extensive manual validation to identify the exact N-terminal position. The situation becomes even more complex when post-translational modifications take place at the neo-N-terminus. Although N-terminal acetylation occurs mostly in the cytosol, it is also observed in some organelles such as chloroplast. To date, no bioinformatics tool is available to define mature protein starting positions, the associated N-terminus acetylation status and/or yield for each proteoform. In this context, we have developed the EnCOUNTer tool (i) to score all characterized peptides using discriminating parameters to identify bona fide mature protein N-termini and (ii) to determine the N-terminus acetylation yield of the most reliable ones.
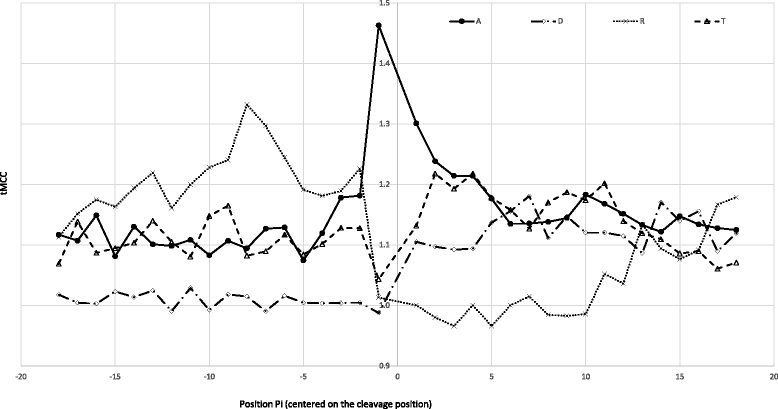
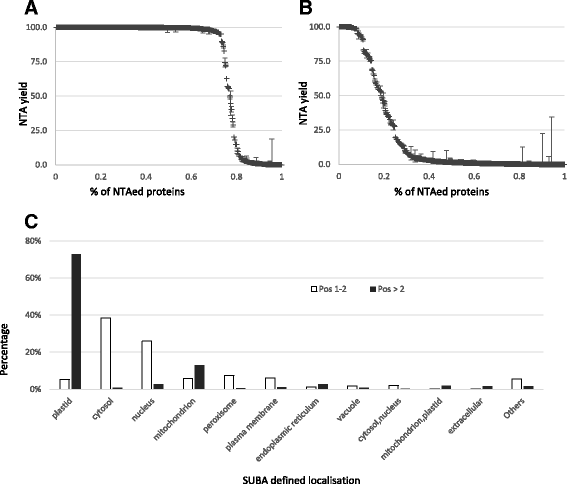
Results: Based on large scale proteomics analyses using the SILProNAQ methodology, tandem mass spectrometry favoured the characterization of thousands of peptides. Data processing using the EnCOUNTer tool provided an efficient and rapid way to extract the most reliable mature protein N-termini. Selected peptides were subjected to N-terminus acetylation yield determination. In an A. thaliana cell lysate, 1232 distinct proteotypic N-termini were characterized of which 648 were located at the predicted protein N-terminus (position 1/2) and 584 were located further downstream (starting at position > 2). A large number of these N-termini were associated with various well-defined maturation processes occurring on organelle-targeted proteins (mitochondria, chloroplast and peroxisome), secreted proteins or membrane-targeted proteins. It was also possible to highlight some protein alternative starts, splicing variants or erroneous protein sequence predictions.
Conclusions: The EnCOUNTer tool provides a unique way to extract accurately the most relevant mature proteins N-terminal peptides from large scale experimental datasets. Such data processing allows the identification of the exact N-terminus position and the associated acetylation yield.
Keywords: Acetylation; Cleavage site; N-terminal modifications; Organelle proteins; Processing tool; Protein maturation; Quantitation; Transit peptide.
Figures
References
-
- Bienvenut WV, Espagne C, Martinez A, Majeran W, Valot B, Zivy M, Vallon O, Adam Z, Meinnel T, Giglione C. Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics. 2011;11(9):1734–1750. doi: 10.1002/pmic.201000634. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

