Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves
- PMID: 28320327
- PMCID: PMC5359985
- DOI: 10.1186/s12870-017-1012-8
Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves
Abstract
Background: As the predominant secondary metabolic pathway in tea plants, flavonoid biosynthesis increases with increasing temperature and illumination. However, the concentration of most flavonoids decreases greatly in light-sensitive tea leaves when they are exposed to light, which further improves tea quality. To reveal the metabolism and potential functions of flavonoids in tea leaves, a natural light-sensitive tea mutant (Huangjinya) cultivated under different light conditions was subjected to metabolomics analysis.
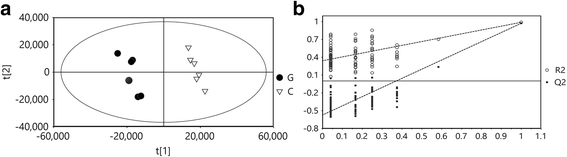
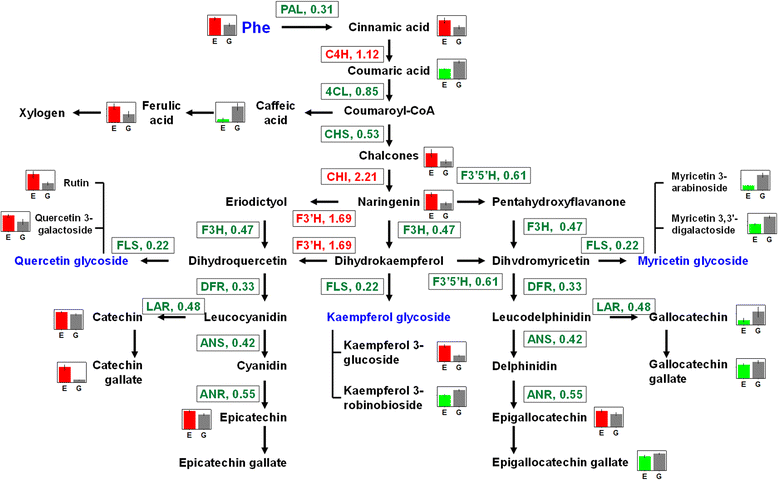
Results: The results showed that chlorotic tea leaves accumulated large amounts of flavonoids with ortho-dihydroxylated B-rings (e.g., catechin gallate, quercetin and its glycosides etc.), whereas total flavonoids (e.g., myricetrin glycoside, epigallocatechin gallate etc.) were considerably reduced, suggesting that the flavonoid components generated from different metabolic branches played different roles in tea leaves. Furthermore, the intracellular localization of flavonoids and the expression pattern of genes involved in secondary metabolic pathways indicate a potential photoprotective function of dihydroxylated flavonoids in light-sensitive tea leaves.
Conclusions: Our results suggest that reactive oxygen species (ROS) scavenging and the antioxidation effects of flavonoids help chlorotic tea plants survive under high light stress, providing new evidence to clarify the functional roles of flavonoids, which accumulate to high levels in tea plants. Moreover, flavonoids with ortho-dihydroxylated B-rings played a greater role in photo-protection to improve the acclimatization of tea plants.
Keywords: Camellia sinensis; Flavonoids; Light sensitive; Metabolism; Photo-protection.
Figures




Similar articles
-
Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves.Molecules. 2022 Sep 7;27(18):5792. doi: 10.3390/molecules27185792. Molecules. 2022. PMID: 36144526 Free PMC article.
-
Dynamic Changes in the Antioxidative Defense System in the Tea Plant Reveal the Photoprotection-Mediated Temporal Accumulation of Flavonoids under Full Sunlight Exposure.Plant Cell Physiol. 2022 Nov 22;63(11):1695-1708. doi: 10.1093/pcp/pcac125. Plant Cell Physiol. 2022. PMID: 36043695
-
Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants (Camellia sinensis) in response to shading.BMC Plant Biol. 2018 Oct 12;18(1):233. doi: 10.1186/s12870-018-1440-0. BMC Plant Biol. 2018. PMID: 30314466 Free PMC article.
-
Exploring plant metabolic genomics: chemical diversity, metabolic complexity in the biosynthesis and transport of specialized metabolites with the tea plant as a model.Crit Rev Biotechnol. 2020 Aug;40(5):667-688. doi: 10.1080/07388551.2020.1752617. Epub 2020 Apr 22. Crit Rev Biotechnol. 2020. PMID: 32321331 Review.
-
Flavonoid metabolites in tea plant (Camellia sinensis) stress response: Insights from bibliometric analysis.Plant Physiol Biochem. 2023 Sep;202:107934. doi: 10.1016/j.plaphy.2023.107934. Epub 2023 Aug 3. Plant Physiol Biochem. 2023. PMID: 37572493 Review.
Cited by
-
Integrative Metabolomic and Transcriptomic Landscape during Akebia trifoliata Fruit Ripening and Cracking.Int J Mol Sci. 2023 Nov 24;24(23):16732. doi: 10.3390/ijms242316732. Int J Mol Sci. 2023. PMID: 38069056 Free PMC article.
-
Characterization of Effects of Different Tea Harvesting Seasons on Quality Components, Color and Sensory Quality of "Yinghong 9" and "Huangyu" Large-Leaf-Variety Black Tea.Molecules. 2022 Dec 9;27(24):8720. doi: 10.3390/molecules27248720. Molecules. 2022. PMID: 36557856 Free PMC article.
-
Transcriptome and metabolome analyses reveal new insights into chlorophyll, photosynthesis, metal ion and phenylpropanoids related pathways during sugarcane ratoon chlorosis.BMC Plant Biol. 2022 Apr 29;22(1):222. doi: 10.1186/s12870-022-03588-8. BMC Plant Biol. 2022. PMID: 35484490 Free PMC article.
-
Transcriptomic and Metabolomic Differences Between Two Saposhnikovia divaricata (Turcz.) Schischk Phenotypes With Single- and Double-Headed Roots.Front Bioeng Biotechnol. 2021 Oct 28;9:764093. doi: 10.3389/fbioe.2021.764093. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34778235 Free PMC article.
-
Investigation of the quality of Lu'an Guapian tea during Grain Rain period by sensory evaluation, objective quantitative indexes and metabolomics.Food Chem X. 2024 Jun 27;23:101595. doi: 10.1016/j.fochx.2024.101595. eCollection 2024 Oct 30. Food Chem X. 2024. PMID: 39071934 Free PMC article.
References
-
- Zhang Q, Ruan J. Tea: analysis and tasting. In: Caballero B, Finglas PM, Toldrá F, editors. Encyclopedia of food and health. Oxford: Academic; 2015. pp. 256–67.
-
- Wang Y, Gao L, Wang Z, Liu Y, Sun M, Yang D, Wei C, Shan Y, Xia T. Light-induced expression of genes involved in phenylpropanoid biosynthetic pathways in callus of tea (Camellia sinensis (L.) O. Kuntze) Sci Hortic. 2012;133:72–83. doi: 10.1016/j.scienta.2011.10.017. - DOI
-
- Zhang Q, Shi Y, Ma L, Yi X, Ruan J. Metabolomic analysis using Ultra-Performance Liquid Chromatography-Quadrupole-Time of Flight Mass Spectrometry (UPLC-Q-TOF/MS) uncovers the effects of light intensity and temperature under shading treatments on the metabolites in tea. PLoS One. 2014;9:e112572. doi: 10.1371/journal.pone.0112572. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

