Co-circulation of West Nile virus and distinct insect-specific flaviviruses in Turkey
- PMID: 28320443
- PMCID: PMC5360070
- DOI: 10.1186/s13071-017-2087-7
Co-circulation of West Nile virus and distinct insect-specific flaviviruses in Turkey
Abstract
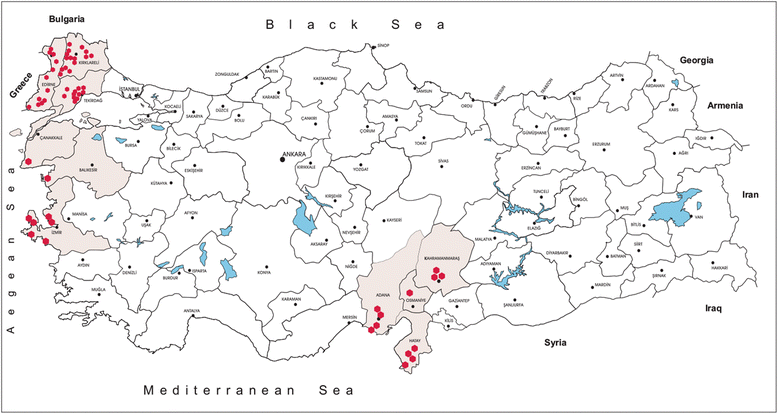
Background: Active vector surveillance provides an efficient tool for monitoring the presence or spread of emerging or re-emerging vector-borne viruses. This study was undertaken to investigate the circulation of flaviviruses. Mosquitoes were collected from 58 locations in 10 provinces across the Aegean, Thrace and Mediterranean Anatolian regions of Turkey in 2014 and 2015. Following morphological identification, mosquitoes were pooled and screened by nested and real-time PCR assays. Detected viruses were further characterised by sequencing. Positive pools were inoculated onto cell lines for virus isolation. Next generation sequencing was employed for genomic characterisation of the isolates.
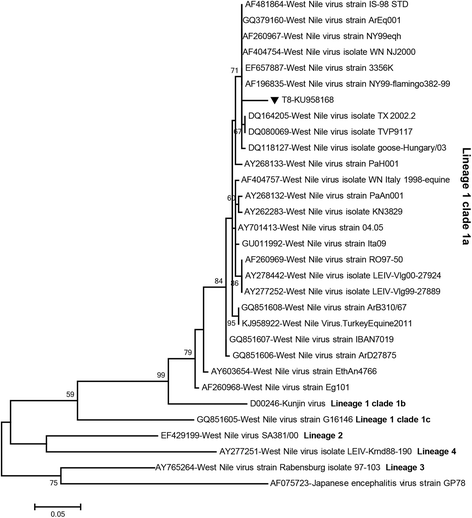
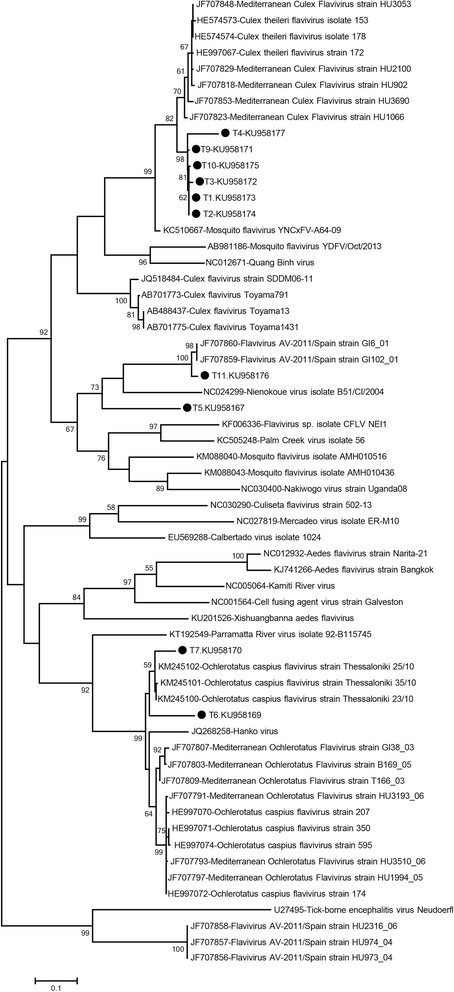
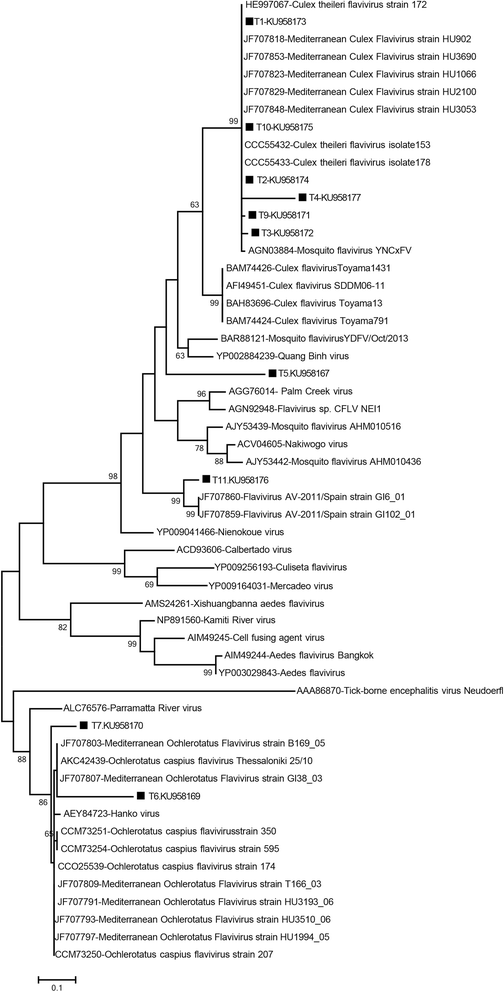
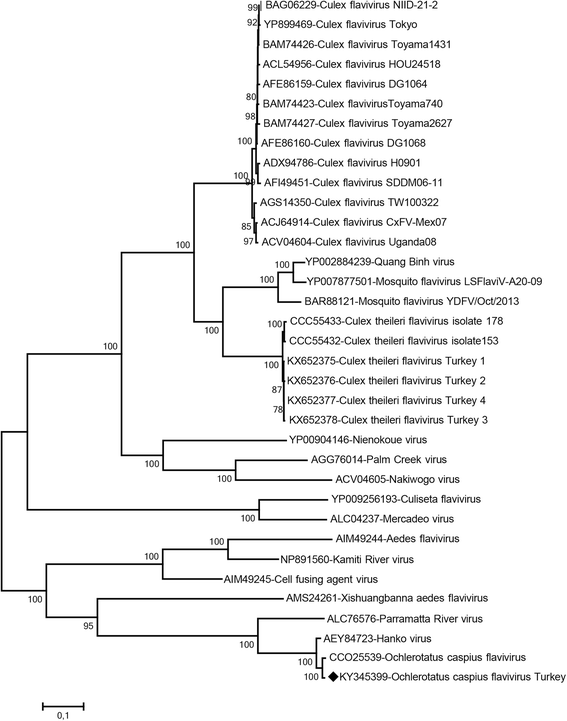
Results: A total of 12,711 mosquito specimens representing 15 species were screened in 594 pools. Eleven pools (2%) were reactive in the virus screening assays. Sequencing revealed West Nile virus (WNV) in one Culex pipiens (s.l.) pool from Thrace. WNV sequence corresponded to lineage one clade 1a but clustered distinctly from the Turkish prototype isolate. In 10 pools, insect-specific flaviviruses were characterised as Culex theileri flavivirus in 5 pools of Culex theileri and one pool of Cx. pipiens (s.l.), Ochlerotatus caspius flavivirus in two pools of Aedes (Ochlerotatus) caspius, Flavivirus AV-2011 in one pool of Culiseta annulata, and an undetermined flavivirus in one pool of Uranotaenia unguiculata from the Aegean and Thrace regions. DNA forms or integration of the detected insect-specific flaviviruses were not observed. A virus strain, tentatively named as "Ochlerotatus caspius flavivirus Turkey", was isolated from an Ae. caspius pool in C6/36 cells. The viral genome comprised 10,370 nucleotides with a putative polyprotein of 3,385 amino acids that follows the canonical flavivirus polyprotein organisation. Sequence comparisons and phylogenetic analyses revealed the close relationship of this strain with Ochlerotatus caspius flavivirus from Portugal and Hanko virus from Finland. Several conserved structural and amino acid motifs were identified.
Conclusions: We identified WNV and several distinct insect-specific flaviviruses during an extensive biosurveillance study of mosquitoes in various regions of Turkey in 2014 and 2015. Ongoing circulation of WNV is revealed, with an unprecedented genetic diversity. A probable replicating form of an insect flavivirus identified only in DNA form was detected.
Keywords: Biosurveillance; Flavivirus; Insect-specific; Mosquito; Turkey; West Nile virus.
Figures
References
-
- Simmonds P, Becher P, Collett MS, Gould EA, Heinz FX, Meyers G, et al. Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2011. Family Flaviviridae. In: Virus taxonomy: classification and nomenclature of viruses; pp. 1003–1020.
-
- Moureau G, Cook S, Lemey P, Nougairede A, Forrester NL, Khasnatinov M, et al. New insights into flavivirus evolution, taxonomy and biogeographic history, extended by analysis of canonical and alternative coding sequences. PLoS One. 2015;10:e0117849. doi: 10.1371/journal.pone.0117849. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

