Pan-cancer analysis distinguishes transcriptional changes of aneuploidy from proliferation
- PMID: 28320919
- PMCID: PMC5378169
- DOI: 10.1101/gr.212225.116
Pan-cancer analysis distinguishes transcriptional changes of aneuploidy from proliferation
Abstract
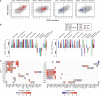
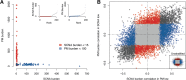
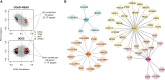
Patterns of gene expression in tumors can arise as a consequence of or result in genomic instability, characterized by the accumulation of somatic copy number alterations (SCNAs) and point mutations (PMs). Expression signatures have been widely used as markers for genomic instability, and both SCNAs and PMs could be thought to associate with distinct signatures given their different formation mechanisms. Here we test this notion by systematically investigating SCNA, PM, and transcriptome data from 2660 cancer patients representing 11 tumor types. Notably, our data indicate that similar expression signatures can be derived from correlating gene expression with either SCNA or PM load. Gene sets related to cell growth and proliferation generally associated positively, and immunoregulatory gene sets negatively, with variant burden. In-depth analyses revealed several genes whose de-regulation correlates with SCNA but not with PM burden, yielding downstream effectors of TP53 and MYC signaling unique to high-SCNA tumors. We compared our findings to expression changes observed in two different cancer mouse models with persistent mitotic chromosomal instability, observing a decrease in proliferative expression signatures. Our results suggest that overexpression of cell-cycle-related genes are a characteristic of proliferation, and likely tumor evolution, rather than ongoing genomic instability.
© 2017 Buccitelli et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
