Prion protein dynamics before aggregation
- PMID: 28320943
- PMCID: PMC5389334
- DOI: 10.1073/pnas.1620400114
Prion protein dynamics before aggregation
Abstract
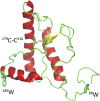
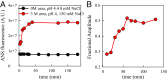
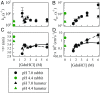
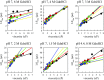
Prion diseases, like Alzheimer's disease and Parkinson disease, are rapidly progressive neurodegenerative disorders caused by misfolding followed by aggregation and accumulation of protein deposits in neuronal cells. Here we measure intramolecular polypeptide backbone reconfiguration as a way to understand the molecular basis of prion aggregation. Our hypothesis is that when reconfiguration is either much faster or much slower than bimolecular diffusion, biomolecular association is not stable, but as the reconfiguration rate becomes similar to the rate of biomolecular diffusion, the association is more stable and subsequent aggregation is faster. Using the technique of Trp-Cys contact quenching, we investigate the effects of various conditions on reconfiguration dynamics of the Syrian hamster and rabbit prion proteins. This protein exhibits behavior in all three reconfiguration regimes. We conclude that the hamster prion is prone to aggregation at pH 4.4 because its reconfiguration rate is slow enough to expose hydrophobic residues on the same time scale that bimolecular association occurs, whereas the rabbit sequence avoids aggregation by reconfiguring 10 times faster than the hamster sequence.
Keywords: astemizole; intramolecular diffusion; prion disease; protein aggregation; protein folding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




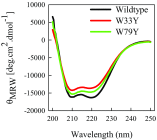

References
-
- Liemann S, Glockshuber R. Transmissible spongiform encephalopathies. Biochem Biophys Res Commun. 1998;250(2):187–193. - PubMed
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–144. - PubMed
-
- Cohen FE, et al. Structural clues to prion replication. Science. 1994;264(5158):530–531. - PubMed
-
- Horwich AL, Weissman JS. Deadly conformations--protein misfolding in prion disease. Cell. 1997;89(4):499–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

