Cerebellar granule cells encode the expectation of reward
- PMID: 28321129
- PMCID: PMC5532014
- DOI: 10.1038/nature21726
Cerebellar granule cells encode the expectation of reward
Abstract
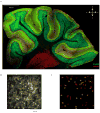
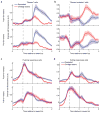
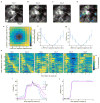
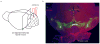
The human brain contains approximately 60 billion cerebellar granule cells, which outnumber all other brain neurons combined. Classical theories posit that a large, diverse population of granule cells allows for highly detailed representations of sensorimotor context, enabling downstream Purkinje cells to sense fine contextual changes. Although evidence suggests a role for the cerebellum in cognition, granule cells are known to encode only sensory and motor context. Here, using two-photon calcium imaging in behaving mice, we show that granule cells convey information about the expectation of reward. Mice initiated voluntary forelimb movements for delayed sugar-water reward. Some granule cells responded preferentially to reward or reward omission, whereas others selectively encoded reward anticipation. Reward responses were not restricted to forelimb movement, as a Pavlovian task evoked similar responses. Compared to predictable rewards, unexpected rewards elicited markedly different granule cell activity despite identical stimuli and licking responses. In both tasks, reward signals were widespread throughout multiple cerebellar lobules. Tracking the same granule cells over several days of learning revealed that cells with reward-anticipating responses emerged from those that responded at the start of learning to reward delivery, whereas reward-omission responses grew stronger as learning progressed. The discovery of predictive, non-sensorimotor encoding in granule cells is a major departure from the current understanding of these neurons and markedly enriches the contextual information available to postsynaptic Purkinje cells, with important implications for cognitive processing in the cerebellum.
Conflict of interest statement
The authors declare no competing financial interests.
Figures









Comment in
-
Cerebellum: The little learning brain.Nat Rev Neurosci. 2017 May;18(5):263. doi: 10.1038/nrn.2017.47. Epub 2017 Apr 6. Nat Rev Neurosci. 2017. PMID: 28381834 No abstract available.
References
-
- Albus JS. A theory of cerebellar function. Math Biosci. 1971;10:25–61.
-
- Fujita M. Adaptive filter model of the cerebellum. Biol Cybern. 1982;45:195–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

