A clear bias in parental origin of de novo pathogenic CNVs related to intellectual disability, developmental delay and multiple congenital anomalies
- PMID: 28322228
- PMCID: PMC5359547
- DOI: 10.1038/srep44446
A clear bias in parental origin of de novo pathogenic CNVs related to intellectual disability, developmental delay and multiple congenital anomalies
Abstract
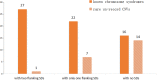
Copy number variation (CNV) is of great significance in human evolution and disorders. Through tracing the parent-of-origin of de novo pathogenic CNVs, we are expected to investigate the relative contributions of germline genomic stability on reproductive health. In our study, short tandem repeat (STR) and single nucleotide polymorphism (SNP) were used to determine the parent-of-origin of 87 de novo pathogenic CNVs found in unrelated patients with intellectual disability (ID), developmental delay (DD) and multiple congenital anomalies (MCA). The results shown that there was a significant difference on the distribution of the parent-of-origin for different CNVs types (Chi-square test, p = 4.914 × 10-3). An apparently paternal bias existed in deletion CNVs and a maternal bias in duplication CNVs, indicating that the relative contribution of paternal germline variations is greater than that of maternal to the origin of deletions, and vice versa to the origin of duplications. By analyzing the sequences flanking the breakpoints, we also confirmed that non-allelic homologous recombination (NAHR) served as the major mechanism for the formation of recurrent CNVs whereas non-SDs-based mechanisms played a part in generating rare non-recurrent CNVs and might relate to the paternal germline bias in deletion CNVs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Sebat J. et al.. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004). - PubMed
-
- Zarrei M., MacDonald J. R., Merico D. & Scherer S. W. A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183 (2015). - PubMed
-
- Feuk L., Carson A. R. & Scherer S. W. Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97 (2006). - PubMed
-
- Weiss L. A. et al.. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 358, 667–675 (2008). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

