Genomic innovations linked to infection strategies across emerging pathogenic chytrid fungi
- PMID: 28322291
- PMCID: PMC5364385
- DOI: 10.1038/ncomms14742
Genomic innovations linked to infection strategies across emerging pathogenic chytrid fungi
Abstract
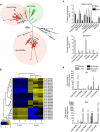
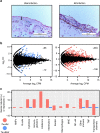
To understand the evolutionary pathways that lead to emerging infections of vertebrates, here we explore the genomic innovations that allow free-living chytrid fungi to adapt to and colonize amphibian hosts. Sequencing and comparing the genomes of two pathogenic species of Batrachochytrium to those of close saprophytic relatives reveals that pathogenicity is associated with remarkable expansions of protease and cell wall gene families, while divergent infection strategies are linked to radiations of lineage-specific gene families. By comparing the host-pathogen response to infection for both pathogens, we illuminate the traits that underpin a strikingly different immune response within a shared host species. Our results show that, despite commonalities that promote infection, specific gene-family radiations contribute to distinct infection strategies. The breadth and evolutionary novelty of candidate virulence factors that we discover underscores the urgent need to halt the advance of pathogenic chytrids and prevent incipient loss of biodiversity.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Fisher M. C., Garner T. W. J. & Walker S. F. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu. Rev. Microbiol. 63, 291–310 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

