Multiplex Substrate Profiling by Mass Spectrometry for Kinases as a Method for Revealing Quantitative Substrate Motifs
- PMID: 28322550
- PMCID: PMC5500290
- DOI: 10.1021/acs.analchem.6b05002
Multiplex Substrate Profiling by Mass Spectrometry for Kinases as a Method for Revealing Quantitative Substrate Motifs
Abstract
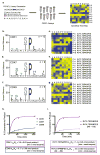
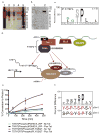
The more than 500 protein kinases comprising the human kinome catalyze hundreds of thousands of phosphorylation events to regulate a diversity of cellular functions; however, the extended substrate specificity is still unknown for many of these kinases. We report here a method for quantitatively describing kinase substrate specificity using an unbiased peptide library-based approach with direct measurement of phosphorylation by tandem liquid chromatography-tandem mass spectrometry (LC-MS/MS) peptide sequencing (multiplex substrate profiling by mass spectrometry, MSP-MS). This method can be deployed with as low as 10 nM enzyme to determine activity against S/T/Y-containing peptides; additionally, label-free quantitation is used to ascertain catalytic efficiency values for individual peptide substrates in the multiplex assay. Using this approach we developed quantitative motifs for a selection of kinases from each branch of the kinome, with and without known substrates, highlighting the applicability of the method. The sensitivity of this approach is evidenced by its ability to detect phosphorylation events from nanogram quantities of immunoprecipitated material, which allows for wider applicability of this method. To increase the information content of the quantitative kinase motifs, a sublibrary approach was used to expand the testable sequence space within a peptide library of approximately 100 members for CDK1, CDK7, and CDK9. Kinetic analysis of the HIV-1 Tat (transactivator of transcription)-positive transcription elongation factor b (P-TEFb) interaction allowed for localization of the P-TEFb phosphorylation site as well as characterization of the stimulatory effect of Tat on P-TEFb catalytic efficiency.
Figures

Similar articles
-
Cyclin-dependent kinase 7 (CDK7)-mediated phosphorylation of the CDK9 activation loop promotes P-TEFb assembly with Tat and proviral HIV reactivation.J Biol Chem. 2018 Jun 29;293(26):10009-10025. doi: 10.1074/jbc.RA117.001347. Epub 2018 May 9. J Biol Chem. 2018. PMID: 29743242 Free PMC article.
-
CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA.Mol Cell Biol. 2000 Sep;20(18):6958-69. doi: 10.1128/MCB.20.18.6958-6969.2000. Mol Cell Biol. 2000. PMID: 10958691 Free PMC article.
-
Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes.PLoS Pathog. 2013;9(5):e1003338. doi: 10.1371/journal.ppat.1003338. Epub 2013 May 2. PLoS Pathog. 2013. PMID: 23658523 Free PMC article. Clinical Trial.
-
The emerging picture of CDK9/P-TEFb: more than 20 years of advances since PITALRE.Mol Biosyst. 2017 Jan 31;13(2):246-276. doi: 10.1039/c6mb00387g. Mol Biosyst. 2017. PMID: 27833949 Review.
-
Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression.J Cell Biochem. 1999 Dec 1;75(3):357-68. J Cell Biochem. 1999. PMID: 10536359 Review.
Cited by
-
High-throughput Identification of FLT3 Wild-type and Mutant Kinase Substrate Preferences and Application to Design of Sensitive In Vitro Kinase Assay Substrates.Mol Cell Proteomics. 2019 Mar;18(3):477-489. doi: 10.1074/mcp.RA118.001111. Epub 2018 Dec 12. Mol Cell Proteomics. 2019. PMID: 30541869 Free PMC article.
-
Accurate Models of Substrate Preferences of Post-Translational Modification Enzymes from a Combination of mRNA Display and Deep Learning.ACS Cent Sci. 2022 Jun 22;8(6):814-824. doi: 10.1021/acscentsci.2c00223. Epub 2022 May 26. ACS Cent Sci. 2022. PMID: 35756369 Free PMC article.
-
Recent Advances in Protein Kinase Activity Analysis Based on Nanomaterials.Int J Mol Sci. 2019 Mar 21;20(6):1440. doi: 10.3390/ijms20061440. Int J Mol Sci. 2019. PMID: 30901923 Free PMC article. Review.
-
Mapping the Protein Kinome: Current Strategy and Future Direction.Cells. 2023 Mar 17;12(6):925. doi: 10.3390/cells12060925. Cells. 2023. PMID: 36980266 Free PMC article. Review.
-
In vitro methods for testing antiviral drugs.Biotechnol Adv. 2018 May-Jun;36(3):557-576. doi: 10.1016/j.biotechadv.2017.12.016. Epub 2017 Dec 29. Biotechnol Adv. 2018. PMID: 29292156 Free PMC article. Review.
References
-
- Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. Chem Rev. 2001;101:2449–2476. - PubMed
-
- Johnson GL, Lapadat R. Science. 2002;298:1911–1912. - PubMed
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. - PubMed
-
- Cohen P. Trends Biochem Sci. 2000;25:596–601. - PubMed
-
- Walsh CT, Garneau-Tsodikova S, Gatto GJ. Angew Chem, Int Ed. 2005;44:7342–7372. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

