Determining Cysteines Available for Covalent Inhibition Across the Human Kinome
- PMID: 28326775
- PMCID: PMC5493210
- DOI: 10.1021/acs.jmedchem.6b01815
Determining Cysteines Available for Covalent Inhibition Across the Human Kinome
Abstract
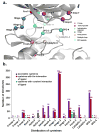
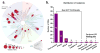
Covalently bound protein kinase inhibitors have been frequently designed to target noncatalytic cysteines at the ATP binding site. Thus, it is important to know if a given cysteine can form a covalent bond. Here we combine a function-site interaction fingerprint method and DFT calculations to determine the potential of cysteines to form a covalent interaction with an inhibitor. By harnessing the human structural kinome, a comprehensive structure-based binding site cysteine data set was assembled. The orientation of the cysteine thiol group indicates which cysteines can potentially form covalent bonds. These covalent inhibitor easy-available cysteines are located within five regions: P-loop, roof of pocket, front pocket, catalytic-2 of the catalytic loop, and DFG-3 close to the DFG peptide. In an independent test set these cysteines covered 95% of covalent kinase inhibitors. This study provides new insights into cysteine reactivity and preference which is important for the prospective development of covalent kinase inhibitors.
Figures





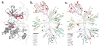

Similar articles
-
Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.Cell Chem Biol. 2019 Jun 20;26(6):818-829.e9. doi: 10.1016/j.chembiol.2019.02.021. Epub 2019 Apr 11. Cell Chem Biol. 2019. PMID: 30982749 Free PMC article.
-
Identification of Covalent Binding Sites Targeting Cysteines Based on Computational Approaches.Mol Pharm. 2016 Sep 6;13(9):3106-18. doi: 10.1021/acs.molpharmaceut.6b00302. Epub 2016 Aug 10. Mol Pharm. 2016. PMID: 27483186
-
Assessing Lysine and Cysteine Reactivities for Designing Targeted Covalent Kinase Inhibitors.J Am Chem Soc. 2019 Apr 24;141(16):6553-6560. doi: 10.1021/jacs.8b13248. Epub 2019 Apr 16. J Am Chem Soc. 2019. PMID: 30945531 Free PMC article.
-
The Cysteinome of Protein Kinases as a Target in Drug Development.Angew Chem Int Ed Engl. 2018 Apr 9;57(16):4372-4385. doi: 10.1002/anie.201707875. Epub 2018 Feb 2. Angew Chem Int Ed Engl. 2018. PMID: 28994500 Review.
-
Targeting protein kinases with selective and semipromiscuous covalent inhibitors.Methods Enzymol. 2014;548:93-116. doi: 10.1016/B978-0-12-397918-6.00004-5. Methods Enzymol. 2014. PMID: 25399643 Free PMC article. Review.
Cited by
-
Reactivity of Covalent Fragments and Their Role in Fragment Based Drug Discovery.Pharmaceuticals (Basel). 2022 Nov 8;15(11):1366. doi: 10.3390/ph15111366. Pharmaceuticals (Basel). 2022. PMID: 36355538 Free PMC article. Review.
-
Reversible covalent c-Jun N-terminal kinase inhibitors targeting a specific cysteine by precision-guided Michael-acceptor warheads.Nat Commun. 2024 Oct 4;15(1):8606. doi: 10.1038/s41467-024-52573-2. Nat Commun. 2024. PMID: 39366946 Free PMC article.
-
Proteome-Wide Profiling of the Covalent-Druggable Cysteines with a Structure-Based Deep Graph Learning Network.Research (Wash D C). 2022 Jul 21;2022:9873564. doi: 10.34133/2022/9873564. eCollection 2022. Research (Wash D C). 2022. PMID: 35958111 Free PMC article.
-
Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.Cell Chem Biol. 2019 Jun 20;26(6):818-829.e9. doi: 10.1016/j.chembiol.2019.02.021. Epub 2019 Apr 11. Cell Chem Biol. 2019. PMID: 30982749 Free PMC article.
-
Structural and biophysical insights into the mode of covalent binding of rationally designed potent BMX inhibitors.RSC Chem Biol. 2020 Aug 28;1(4):251-262. doi: 10.1039/d0cb00033g. eCollection 2020 Oct 1. RSC Chem Biol. 2020. PMID: 34458764 Free PMC article.
References
-
- Lahiry P, Torkamani A, Schork NJ, Hegele RA. Kinase mutations in human disease: Interpreting genotype-phenotype relationships. Nat Rev Genet. 2010;11:60–74. - PubMed
-
- Muller S, Chaikuad A, Gray NS, Knapp S. The ins and outs of selective kinase inhibitor development. Nat Chem Biol. 2015;11:818–821. - PubMed
-
- Gharwan H, Groninger H. Kinase inhibitors and monoclonal antibodies in oncology: Clinical implications. Nat Rev Clin Oncol. 2016;13:209–227. - PubMed
-
- Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101:2271–2290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

