A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs
- PMID: 28327084
- PMCID: PMC5361837
- DOI: 10.1186/s12864-017-3639-0
A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs
Abstract
Background: Animal's efficiency in converting feed into lean gain is a critical issue for the profitability of meat industries. This study aimed to describe shared and specific molecular responses in different tissues of pigs divergently selected over eight generations for residual feed intake (RFI).
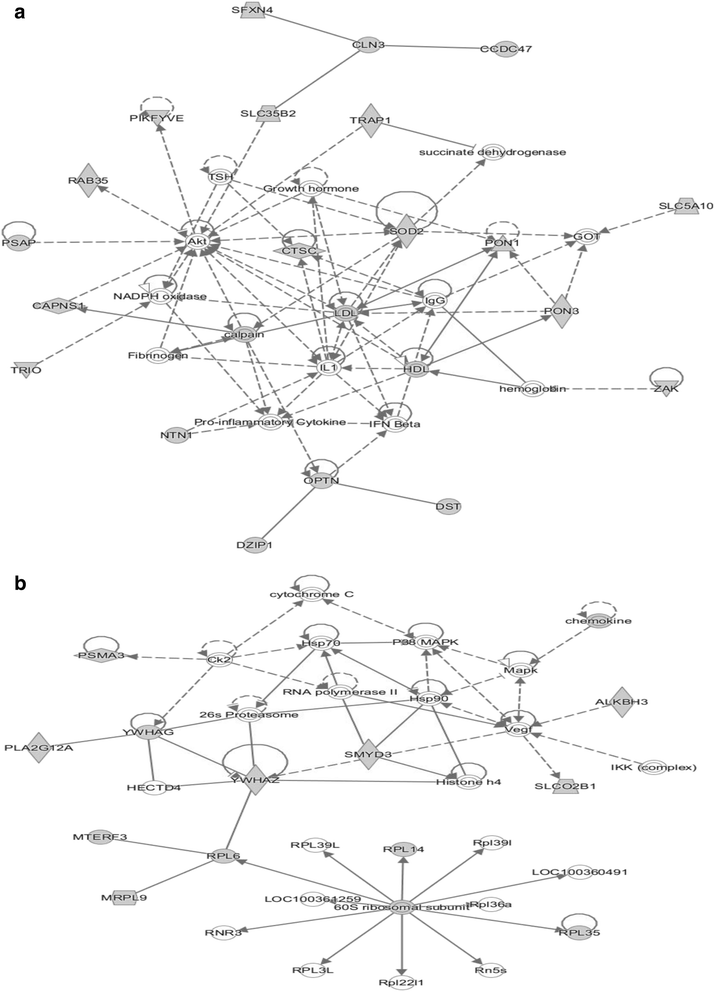
Results: Pigs from the low RFI line had an improved gain-to-feed ratio during the test period and displayed higher leanness but similar adiposity when compared with pigs from the high RFI line at 132 days of age. Transcriptomics data were generated from longissimus muscle, liver and two adipose tissues using a porcine microarray and analyzed for the line effect (n = 24 pigs per line). The most apparent effect of the line was seen in muscle, whereas subcutaneous adipose tissue was the less affected tissue. Molecular data were analyzed by bioinformatics and subjected to multidimensional statistics to identify common biological processes across tissues and key genes participating to differences in the genetics of feed efficiency. Immune response, response to oxidative stress and protein metabolism were the main biological pathways shared by the four tissues that distinguished pigs from the low or high RFI lines. Many immune genes were under-expressed in the four tissues of the most efficient pigs. The main genes contributing to difference between pigs from the low vs high RFI lines were CD40, CTSC and NTN1. Different genes associated with energy use were modulated in a tissue-specific manner between the two lines. The gene expression program related to glycogen utilization was specifically up-regulated in muscle of pigs from the low RFI line (more efficient). Genes involved in fatty acid oxidation were down-regulated in muscle but were promoted in adipose tissues of the same pigs when compared with pigs from the high RFI line (less efficient). This underlined opposite line-associated strategies for energy use in skeletal muscle and adipose tissue. Genes related to cholesterol synthesis and efflux in liver and perirenal fat were also differentially regulated in pigs from the low vs high RFI lines.
Conclusions: Non-productive functions such as immunity, defense against pathogens and oxidative stress contribute likely to inter-individual variations in feed efficiency.
Keywords: Feed efficiency; Multi-tissues; Multiple factor analysis; Pig; Residual feed intake; Transcriptome.
Figures




References
-
- Ifip. Résultats Porc Bretagne. 2016. http://ifip.asso.fr/PagesStatics/resultat/pdf/resultat_bretagne.pdf. Accessed 31 Nov 2016.
-
- Koch RM, Swiger LA, Chambers D, Gregory KE. Efficiency of feed use in beef cattle. J Anim Sci. 1963;22:486–494. doi: 10.2527/jas1963.222486x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

