TRIM8 restores p53 tumour suppressor function by blunting N-MYC activity in chemo-resistant tumours
- PMID: 28327152
- PMCID: PMC5359838
- DOI: 10.1186/s12943-017-0634-7
TRIM8 restores p53 tumour suppressor function by blunting N-MYC activity in chemo-resistant tumours
Abstract
Background: TRIM8 plays a key role in controlling the p53 molecular switch that sustains the transcriptional activation of cell cycle arrest genes and response to chemotherapeutic drugs. The mechanisms that regulate TRIM8, especially in cancers like clear cell Renal Cell Carcinoma (ccRCC) and colorectal cancer (CRC) where it is low expressed, are still unknown. However, recent studies suggest the potential involvement of some microRNAs belonging to miR-17-92 and its paralogous clusters, which could include TRIM8 in a more complex pathway.
Methods: We used RCC and CRC cell models for in-vitro experiments, and ccRCC patients and xenograft transplanted mice for in vivo assessments. To measure microRNAs levels we performed RT-qPCR, while steady-states of TRIM8, p53, p21 and N-MYC were quantified at protein level by Western Blotting as well as at transcript level by RT-qPCR. Luciferase reporter assays were performed to assess the interaction between TRIM8 and specific miRNAs, and the potential effects of this interaction on TRIM8 expression. Moreover, we treated our cell models with conventional chemotherapeutic drugs or tyrosine kinase inhibitors, and measured their response in terms of cell proliferation by MTT and colony suppression assays.
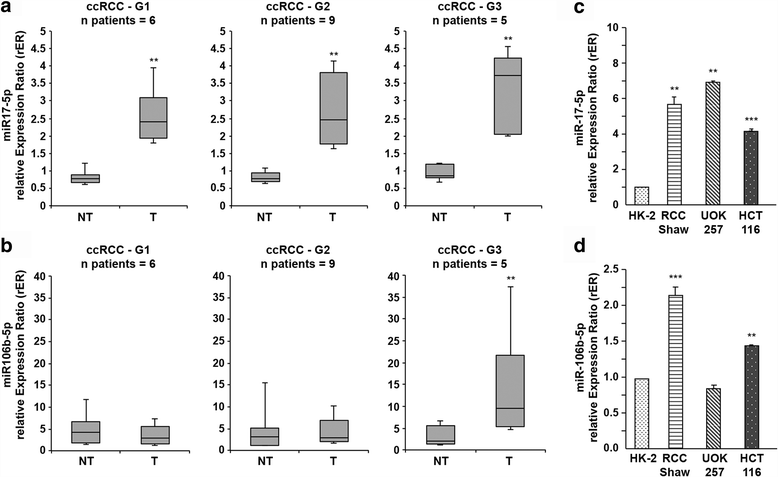
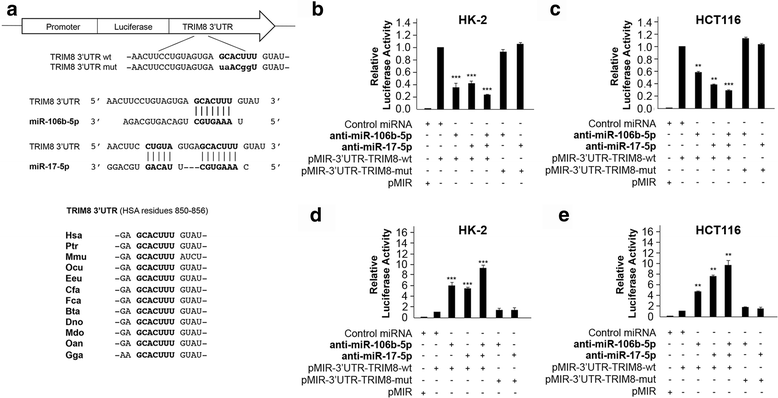
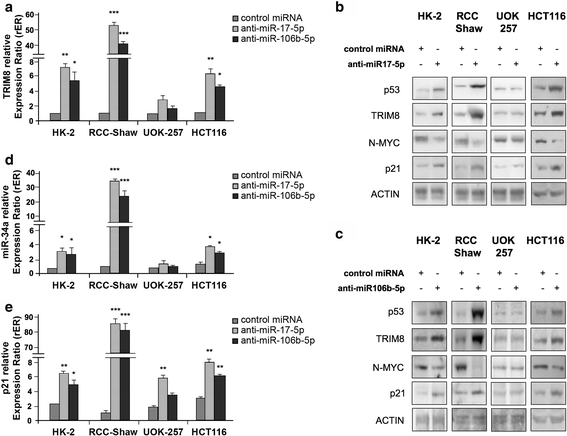
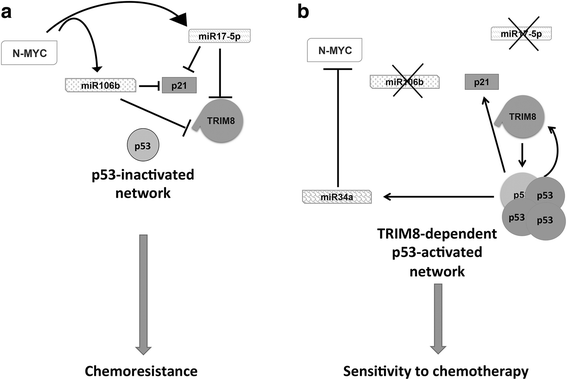
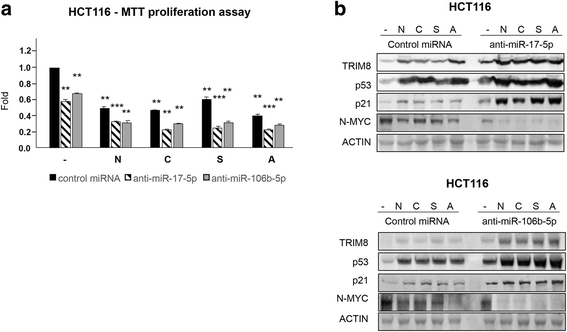
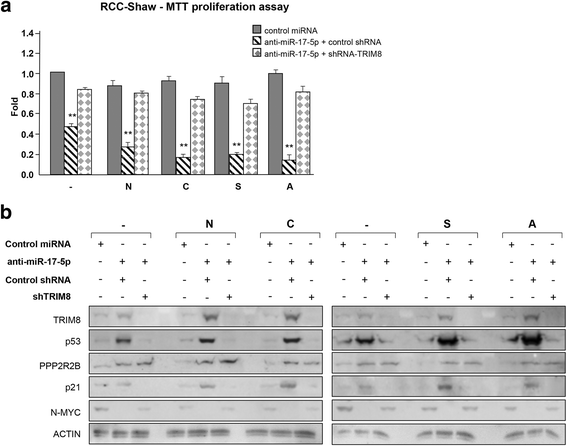
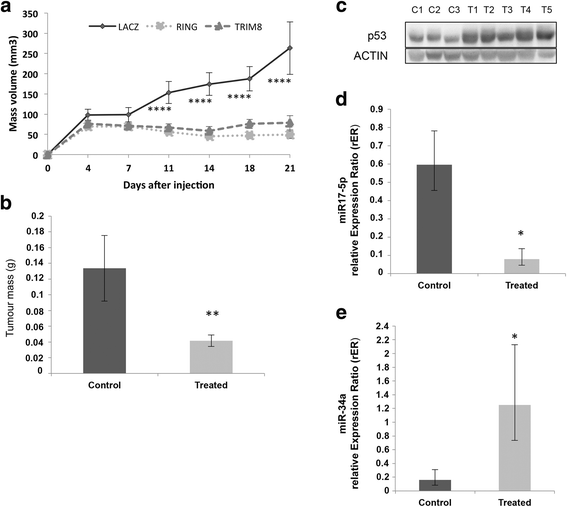
Results: We showed that TRIM8 is a target of miR-17-5p and miR-106b-5p, whose expression is promoted by N-MYC, and that alterations of their levels affect cell proliferation, acting on the TRIM8 transcripts stability, as confirmed in ccRCC patients and cell lines. In addition, reducing the levels of miR-17-5p/miR-106b-5p, we increased the chemo-sensitivity of RCC/CRC-derived cells to anti-tumour drugs used in the clinic. Intriguingly, this occurs, on one hand, by recovering the p53 tumour suppressor activity in a TRIM8-dependent fashion and, on the other hand, by promoting the transcription of miR-34a that turns off the oncogenic action of N-MYC. This ultimately leads to cell proliferation reduction or block, observed also in colon cancer xenografts overexpressing TRIM8.
Conclusions: In this paper we provided evidence that TRIM8 and its regulators miR-17-5p and miR-106b-5 participate to a feedback loop controlling cell proliferation through the reciprocal modulation of p53, miR-34a and N-MYC. Our experiments pointed out that this axis is pivotal in defining drug responsiveness of cancers such ccRCC and CRC.
Keywords: Drug resistance; N-MYC; TRIM8; miR-17 family; p53.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

