Myeloid C-Type Lectin Receptors in Viral Recognition and Antiviral Immunity
- PMID: 28327518
- PMCID: PMC5371814
- DOI: 10.3390/v9030059
Myeloid C-Type Lectin Receptors in Viral Recognition and Antiviral Immunity
Abstract
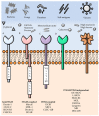
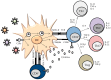
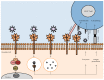
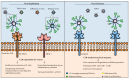
Recognition of viral glycans by pattern recognition receptors (PRRs) in innate immunity contributes to antiviral immune responses. C-type lectin receptors (CLRs) are PRRs capable of sensing glycans present in viral pathogens to activate antiviral immune responses such as phagocytosis, antigen processing and presentation, and subsequent T cell activation. The ability of CLRs to elicit and shape adaptive immunity plays a critical role in the inhibition of viral spread within the host. However, certain viruses exploit CLRs for viral entry into host cells to avoid immune recognition. To block CLR interactions with viral glycoproteins, antiviral strategies may involve the use of multivalent glycan carrier systems. In this review, we describe the role of CLRs in antiviral immunity and we highlight their dual function in viral clearance and exploitation by viral pathogens.
Keywords: C-type lectin receptors; antiviral immunity; dendritic cells; glycans; immunomodulation; macrophages; viral evasion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Murphy K., Travers P., Walport M., Janeway C. Janeway’s Immunobiology. Garland Science; New York, NY, USA: 2012.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

