Quantitative volumetric Raman imaging of three dimensional cell cultures
- PMID: 28327660
- PMCID: PMC5364421
- DOI: 10.1038/ncomms14843
Quantitative volumetric Raman imaging of three dimensional cell cultures
Abstract
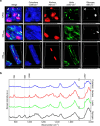
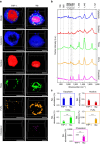
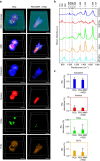
The ability to simultaneously image multiple biomolecules in biologically relevant three-dimensional (3D) cell culture environments would contribute greatly to the understanding of complex cellular mechanisms and cell-material interactions. Here, we present a computational framework for label-free quantitative volumetric Raman imaging (qVRI). We apply qVRI to a selection of biological systems: human pluripotent stem cells with their cardiac derivatives, monocytes and monocyte-derived macrophages in conventional cell culture systems and mesenchymal stem cells inside biomimetic hydrogels that supplied a 3D cell culture environment. We demonstrate visualization and quantification of fine details in cell shape, cytoplasm, nucleus, lipid bodies and cytoskeletal structures in 3D with unprecedented biomolecular specificity for vibrational microspectroscopy.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Ravi M., Paramesh V., Kaviya S. R., Anuradha E. & Paul Solomon F. D. 3D cell culture systems: advantages and applications. J. Cell. Physiol. 230, 16–26 (2015). - PubMed
-
- Justice B. A., Badr N. A. & Felder R. A. 3D cell culture opens new dimensions in cell-based assays. Drug Discov. Today 14, 102–107 (2009). - PubMed
-
- Streets A. M., Li A., Chen T. & Huang Y. Imaging without fluorescence: nonlinear optical microscopy for quantitative cellular imaging. Anal. Chem. 86, 8506–8513 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

