Estimating the total number of phosphoproteins and phosphorylation sites in eukaryotic proteomes
- PMID: 28327990
- PMCID: PMC5466708
- DOI: 10.1093/gigascience/giw015
Estimating the total number of phosphoproteins and phosphorylation sites in eukaryotic proteomes
Abstract
Background: Phosphorylation is the most frequent post-translational modification made to proteins and may regulate protein activity as either a molecular digital switch or a rheostat. Despite the cornucopia of high-throughput (HTP) phosphoproteomic data in the last decade, it remains unclear how many proteins are phosphorylated and how many phosphorylation sites (p-sites) can exist in total within a eukaryotic proteome. We present the first reliable estimates of the total number of phosphoproteins and p-sites for four eukaryotes (human, mouse, Arabidopsis, and yeast).
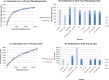
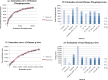
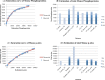
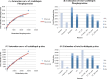
Results: In all, 187 HTP phosphoproteomic datasets were filtered, compiled, and studied along with two low-throughput (LTP) compendia. Estimates of the number of phosphoproteins and p-sites were inferred by two methods: Capture-Recapture, and fitting the saturation curve of cumulative redundant vs. cumulative non-redundant phosphoproteins/p-sites. Estimates were also adjusted for different levels of noise within the individual datasets and other confounding factors. We estimate that in total, 13 000, 11 000, and 3000 phosphoproteins and 230 000, 156 000, and 40 000 p-sites exist in human, mouse, and yeast, respectively, whereas estimates for Arabidopsis were not as reliable.
Conclusions: Most of the phosphoproteins have been discovered for human, mouse, and yeast, while the dataset for Arabidopsis is still far from complete. The datasets for p-sites are not as close to saturation as those for phosphoproteins. Integration of the LTP data suggests that current HTP phosphoproteomics appears to be capable of capturing 70 % to 95 % of total phosphoproteins, but only 40 % to 60 % of total p-sites.
Keywords: Arabidopsis; Capture-Recapture; Curve-Fitting; Phosphoproteomics; human; mouse; total number of phosphoproteins; total number of phosphorylation sites; yeast.
© The Author 2017. Published by Oxford University Press.
Figures
References
-
- Krüger R, Kübler D, Pallissé R et al. . Protein and proteome phosphorylation stoichiometry analysis by element mass spectrometry. Anal. Chem. 2006;78:1987–1994. - PubMed
-
- Cohen P. The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem. Sci. 2000;25:596–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

