Versatile modes of cellular regulation via cyclic dinucleotides
- PMID: 28328921
- PMCID: PMC5773344
- DOI: 10.1038/nchembio.2337
Versatile modes of cellular regulation via cyclic dinucleotides
Abstract
Since the discovery of c-di-GMP almost three decades ago, cyclic dinucleotides (CDNs) have emerged as widely used signaling molecules in most kingdoms of life. The family of second messengers now includes c-di-AMP and distinct versions of mixed cyclic GMP-AMP (cGAMP) compounds. In addition to these nucleotides, a vast number of proteins for the production and turnover of these molecules have been described, as well as effectors that translate the signals into physiological responses. The latter include, but are not limited to, mechanisms for adaptation and survival in prokaryotes, persistence and virulence of bacterial pathogens, and immune responses to viral and bacterial invasion in eukaryotes. In this review, we will focus on recent discoveries and emerging themes that illustrate the ubiquity and versatility of cyclic dinucleotide function at the transcriptional and post-translational levels and, in particular, on insights gained through mechanistic structure-function analyses.
Conflict of interest statement
Figures





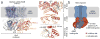
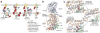

References
-
- Ross P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. Seminal work on the discovery of c-di-GMP as an allosteric regulator of bacterial cellulose synthesis that laid the foundation of CDN signaling research. - PubMed
-
- Weinhouse H, et al. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–211. - PubMed
-
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

