The SWI/SNF Protein PBRM1 Restrains VHL-Loss-Driven Clear Cell Renal Cell Carcinoma
- PMID: 28329682
- PMCID: PMC5431084
- DOI: 10.1016/j.celrep.2017.02.074
The SWI/SNF Protein PBRM1 Restrains VHL-Loss-Driven Clear Cell Renal Cell Carcinoma
Abstract
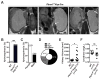
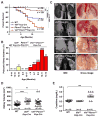
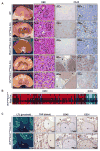
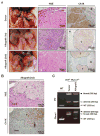
PBRM1 is the second most commonly mutated gene after VHL in clear cell renal cell carcinoma (ccRCC). However, the biological consequences of PBRM1 mutations for kidney tumorigenesis are unknown. Here, we find that kidney-specific deletion of Vhl and Pbrm1, but not either gene alone, results in bilateral, multifocal, transplantable clear cell kidney cancers. PBRM1 loss amplified the transcriptional outputs of HIF1 and STAT3 incurred by Vhl deficiency. Analysis of mouse and human ccRCC revealed convergence on mTOR activation, representing the third driver event after genetic inactivation of VHL and PBRM1. Our study reports a physiological preclinical ccRCC mouse model that recapitulates somatic mutations in human ccRCC and provides mechanistic and therapeutic insights into PBRM1 mutated subtypes of human ccRCC.
Keywords: HIF1; MTOR; PBRM1; STAT3; VHL; ccRCC; epigenetics; genetics; kidney cancer; mouse tumor model.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. - PMC - PubMed
-
- Baba M, Furihata M, Hong SB, Tessarollo L, Haines DC, Southon E, Patel V, Igarashi P, Alvord WG, Leighty R, et al. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst. 2008;100:140–154. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

