Demethylated HSATII DNA and HSATII RNA Foci Sequester PRC1 and MeCP2 into Cancer-Specific Nuclear Bodies
- PMID: 28329686
- PMCID: PMC5429029
- DOI: 10.1016/j.celrep.2017.02.072
Demethylated HSATII DNA and HSATII RNA Foci Sequester PRC1 and MeCP2 into Cancer-Specific Nuclear Bodies
Abstract
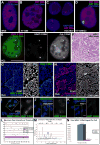
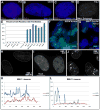
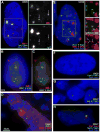
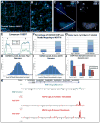
This study reveals that high-copy satellite II (HSATII) sequences in the human genome can bind and impact distribution of chromatin regulatory proteins and that this goes awry in cancer. In many cancers, master regulatory proteins form two types of cancer-specific nuclear bodies, caused by locus-specific deregulation of HSATII. DNA demethylation at the 1q12 mega-satellite, common in cancer, causes PRC1 aggregation into prominent Cancer-Associated Polycomb (CAP) bodies. These loci remain silent, whereas HSATII loci with reduced PRC1 become derepressed, reflecting imbalanced distribution of UbH2A on these and other PcG-regulated loci. Large nuclear foci of HSATII RNA form and sequester copious MeCP2 into Cancer-Associated Satellite Transcript (CAST) bodies. Hence, HSATII DNA and RNA have an exceptional capacity to act as molecular sponges and sequester chromatin regulatory proteins into abnormal nuclear bodies in cancer. The compartmentalization of regulatory proteins within nuclear structure, triggered by demethylation of "junk" repeats, raises the possibility that this contributes to further compromise of the epigenome and neoplastic progression.
Keywords: DNA methylation; breast cancer; cancer biomarkers; cancer epigenetics; centromere; nuclear structure; polycomb proteins; satellite heterochromatin.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bantignies F, Cavalli G. Polycomb group proteins: repression in 3D. Trends Genet. 2011;27:454–464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

