The IFN-λ-IFN-λR1-IL-10Rβ Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity
- PMID: 28329704
- PMCID: PMC5510750
- DOI: 10.1016/j.immuni.2017.02.017
The IFN-λ-IFN-λR1-IL-10Rβ Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity
Abstract
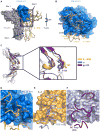
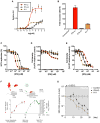
Type III interferons (IFN-λs) signal through a heterodimeric receptor complex composed of the IFN-λR1 subunit, specific for IFN-λs, and interleukin-10Rβ (IL-10Rβ), which is shared by multiple cytokines in the IL-10 superfamily. Low affinity of IL-10Rβ for cytokines has impeded efforts aimed at crystallizing cytokine-receptor complexes. We used yeast surface display to engineer a higher-affinity IFN-λ variant, H11, which enabled crystallization of the ternary complex. The structure revealed that IL-10Rβ uses a network of tyrosine residues as hydrophobic anchor points to engage IL-10 family cytokines that present complementary hydrophobic binding patches, explaining its role as both a cross-reactive but cytokine-specific receptor. H11 elicited increased anti-proliferative and antiviral activities in vitro and in vivo. In contrast, engineered higher-affinity type I IFNs did not increase antiviral potency over wild-type type I IFNs. Our findings provide insight into cytokine recognition by the IL-10R family and highlight the plasticity of type III interferon signaling and its therapeutic potential.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- Andrus L, Marukian S, Jones CT, Catanese MT, Sheahan TP, Schoggins JW, Barry WT, Dustin LB, Trehan K, Ploss A, et al. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

