The recombinant pea defensin Drr230a is active against impacting soybean and cotton pathogenic fungi from the genera Fusarium, Colletotrichum and Phakopsora
- PMID: 28330129
- PMCID: PMC4752952
- DOI: 10.1007/s13205-015-0320-7
The recombinant pea defensin Drr230a is active against impacting soybean and cotton pathogenic fungi from the genera Fusarium, Colletotrichum and Phakopsora
Abstract
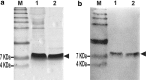
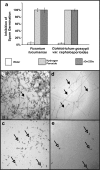
Plant defensins are antifungal peptides produced by the innate immune system plants developed to circumvent fungal infection. The defensin Drr230a, originally isolated from pea, has been previously shown to be active against various entomopathogenic and phytopathogenic fungi. In the present study, the activity of a yeast-expressed recombinant Drr230a protein (rDrr230a) was tested against impacting soybean and cotton fungi. First, the gene was subcloned into the yeast expression vector pPICZαA and expressed in Pichia pastoris. Resulting rDrr230a exhibited in vitro activity against fungal growth and spore germination of Fusarium tucumaniae, which causes soybean sudden death syndrome, and against Colletotrichum gossypii var. cephalosporioides, which causes cotton ramulosis. The rDrr230a IC50 corresponding to inhibition of fungal growth of F. tucumaniae and C. gossypii var. cephalosporioides was 7.67 and 0.84 µM, respectively, demonstrating moderate activity against F. tucumaniae and high potency against C. gossypii var. cephalosporioides. Additionally, rDrr230a at 25 ng/µl (3.83 µM) resulted in 100 % inhibition of spore germination of both fungi, demonstrating that rDrr230a affects fungal development since spore germination. Moreover, rDrr230a at 3 µg/µl (460.12 µM) inhibited 100 % of in vitro spore germination of the obligatory biotrophic fungus Phakopsora pachyrhizi, which causes Asian soybean rust. Interestingly, rDrr230a substantially decreased the severity of Asian rust, as demonstrated by in planta assay. To our knowledge, this is the first report of a plant defensin active against an obligatory biotrophic phytopathogenic fungus. Results revealed the potential of rDrr230a as a candidate to be used in plant genetic engineering to control relevant cotton and soybean fungal diseases.
Keywords: Colletotrichum gossypii var. cephalosporioides; Defensin; Fusarium tucumaniae; Phakopsora pachyrhizi; Pichia pastoris; Pisum sativum.
Conflict of interest statement
The authors declare no financial or commercial conflict of interest.
Figures





Similar articles
-
Field Resistance to Phakopsora pachyrhizi and Colletotrichum truncatum of Transgenic Soybean Expressing the NmDef02 Plant Defensin Gene.Front Plant Sci. 2020 May 26;11:562. doi: 10.3389/fpls.2020.00562. eCollection 2020. Front Plant Sci. 2020. PMID: 32528487 Free PMC article.
-
Endophytic Fungi Inoculation Reduces Ramulosis Severity in Gossypium hirsutum Plants.Microorganisms. 2024 May 31;12(6):1124. doi: 10.3390/microorganisms12061124. Microorganisms. 2024. PMID: 38930506 Free PMC article.
-
A new chitinase-like xylanase inhibitor protein (XIP) from coffee (Coffea arabica) affects Soybean Asian rust (Phakopsora pachyrhizi) spore germination.BMC Biotechnol. 2011 Feb 7;11:14. doi: 10.1186/1472-6750-11-14. BMC Biotechnol. 2011. PMID: 21299880 Free PMC article.
-
The Top 10 fungal pathogens in molecular plant pathology.Mol Plant Pathol. 2012 May;13(4):414-30. doi: 10.1111/j.1364-3703.2011.00783.x. Mol Plant Pathol. 2012. PMID: 22471698 Free PMC article. Review.
-
Mycoherbicides for the Noxious Meddlesome: Can Colletotrichum be a Budding Candidate?Front Microbiol. 2021 Sep 30;12:754048. doi: 10.3389/fmicb.2021.754048. eCollection 2021. Front Microbiol. 2021. PMID: 34659190 Free PMC article. Review.
Cited by
-
The Dual Role of Antimicrobial Proteins and Peptides: Exploring Their Direct Impact and Plant Defense-Enhancing Abilities.Plants (Basel). 2024 Jul 26;13(15):2059. doi: 10.3390/plants13152059. Plants (Basel). 2024. PMID: 39124177 Free PMC article. Review.
-
Field Resistance to Phakopsora pachyrhizi and Colletotrichum truncatum of Transgenic Soybean Expressing the NmDef02 Plant Defensin Gene.Front Plant Sci. 2020 May 26;11:562. doi: 10.3389/fpls.2020.00562. eCollection 2020. Front Plant Sci. 2020. PMID: 32528487 Free PMC article.
-
The Versatile Roles of Sulfur-Containing Biomolecules in Plant Defense-A Road to Disease Resistance.Plants (Basel). 2020 Dec 3;9(12):1705. doi: 10.3390/plants9121705. Plants (Basel). 2020. PMID: 33287437 Free PMC article. Review.
References
-
- Aoki T, O’donell K, Homma Y, Lattanzi AR. Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex—F. virguliforme in North America and F. tucumaniae in South America. Mycologia. 2003;95:660–684. doi: 10.2307/3761942. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

