In silico characterization and differential expression pattern analysis of conserved HMG CoA reductase domain isolated from Aconitum balfourii Stapf
- PMID: 28330159
- PMCID: PMC4781813
- DOI: 10.1007/s13205-016-0405-y
In silico characterization and differential expression pattern analysis of conserved HMG CoA reductase domain isolated from Aconitum balfourii Stapf
Abstract
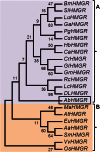
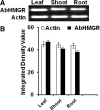
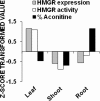
The 3-hydroxy-3-methyl glutaryl CoA reductase (HMGR) is the key enzyme of mevalonate pathway in plants. A partial genomic DNA fragment encoding HMGR conserved domain (denoted as AbHMGR) is isolated from Aconitum balfourii Stapf. It comprises 871 bp encoding 290 amino acids. In silico analysis reveals that it had extensive similarities to other plant HMGR gene. Domain analysis of AbHMGR showed two highly conserved NADPH and HMG CoA domains. Docking study predicted inhibitor, substrate and cofactor binding sites in the protein. Expression analysis revealed that AbHMGR is similarly expressed in all tested tissues with differential pattern. The highest expression was found in leaf tissue. However, fold expression in root and shoot tissue was almost similar. Enzyme activity of HMGR was found to be much higher in leaf tissue as compared to other tissues. The highest aconitine content (0.015 %) was obtained in root tissues. Our data laid a foundation for further investigation of HMGR role in Aconitum balfourii.
Keywords: 3-Hydroxy-3-methyl glutaryl-CoA coenzyme A reductase; Aconitine; Expression profiling; HMGR; Mevalonate pathway.
Conflict of interest statement
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

