Stromal Gene Expression is Predictive for Metastatic Primary Prostate Cancer
- PMID: 28330676
- PMCID: PMC6685211
- DOI: 10.1016/j.eururo.2017.02.038
Stromal Gene Expression is Predictive for Metastatic Primary Prostate Cancer
Abstract
Background: Clinical grading systems using clinical features alongside nomograms lack precision in guiding treatment decisions in prostate cancer (PCa). There is a critical need for identification of biomarkers that can more accurately stratify patients with primary PCa.
Objective: To identify a robust prognostic signature to better distinguish indolent from aggressive prostate cancer (PCa).
Design, setting, and participants: To develop the signature, whole-genome and whole-transcriptome sequencing was conducted on five PCa patient-derived xenograft (PDX) models collected from independent foci of a single primary tumor and exhibiting variable metastatic phenotypes. Multiple independent clinical cohorts including an intermediate-risk cohort were used to validate the biomarkers.
Outcome measurements and statistical analysis: The outcome measurement defining aggressive PCa was metastasis following radical prostatectomy. A generalized linear model with lasso regularization was used to build a 93-gene stroma-derived metastasis signature (SDMS). The SDMS association with metastasis was assessed using a Wilcoxon rank-sum test. Performance was evaluated using the area under the curve (AUC) for the receiver operating characteristic, and Kaplan-Meier curves. Univariable and multivariable regression models were used to compare the SDMS alongside clinicopathological variables and reported signatures. AUC was assessed to determine if SDMS is additive or synergistic to previously reported signatures.
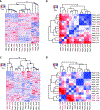
Results and limitations: A close association between stromal gene expression and metastatic phenotype was observed. Accordingly, the SDMS was modeled and validated in multiple independent clinical cohorts. Patients with higher SDMS scores were found to have worse prognosis. Furthermore, SDMS was an independent prognostic factor, can stratify risk in intermediate-risk PCa, and can improve the performance of other previously reported signatures.
Conclusions: Profiling of stromal gene expression led to development of an SDMS that was validated as independently prognostic for the metastatic potential of prostate tumors.
Patient summary: Our stroma-derived metastasis signature can predict the metastatic potential of early stage disease and will strengthen decisions regarding selection of active surveillance versus surgery and/or radiation therapy for prostate cancer patients. Furthermore, profiling of stroma cells should be more consistent than profiling of diverse cellular populations of heterogeneous tumors.
Keywords: Genomic profiling; Prognostic biomarkers; Prostate cancer metastasis; RNA sequencing; Stromal gene.
Copyright © 2017 European Association of Urology. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures



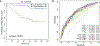
Comment in
-
A Partner in Crime: Tumor-associated Stroma and Metastatic Prostate Cancer.Eur Urol. 2018 Apr;73(4):533-534. doi: 10.1016/j.eururo.2017.11.029. Epub 2017 Dec 22. Eur Urol. 2018. PMID: 29275835 No abstract available.
References
-
- Daskivich TJ, Chamie K, Kwan L, et al. Overtreatment of men with low-risk prostate cancer and significant comorbidity. Cancer 2011;117:2058–66. - PubMed
-
- Lalonde E, Ishkanian AS, Sykes J, et al. Tumour genomic and micro-environmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol 2014;15:1521–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

