Gorilla MHC class I gene and sequence variation in a comparative context
- PMID: 28332079
- PMCID: PMC5400801
- DOI: 10.1007/s00251-017-0974-x
Gorilla MHC class I gene and sequence variation in a comparative context
Abstract
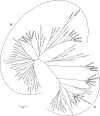
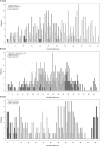
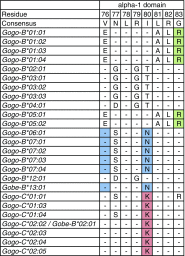
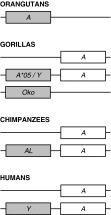
Comparisons of MHC gene content and diversity among closely related species can provide insights into the evolutionary mechanisms shaping immune system variation. After chimpanzees and bonobos, gorillas are humans' closest living relatives; but in contrast, relatively little is known about the structure and variation of gorilla MHC class I genes (Gogo). Here, we combined long-range amplifications and long-read sequencing technology to analyze full-length MHC class I genes in 35 gorillas. We obtained 50 full-length genomic sequences corresponding to 15 Gogo-A alleles, 4 Gogo-Oko alleles, 21 Gogo-B alleles, and 10 Gogo-C alleles including 19 novel coding region sequences. We identified two previously undetected MHC class I genes related to Gogo-A and Gogo-B, respectively, thereby illustrating the potential of this approach for efficient and highly accurate MHC genotyping. Consistent with their phylogenetic position within the hominid family, individual gorilla MHC haplotypes share characteristics with humans and chimpanzees as well as orangutans suggesting a complex history of the MHC class I genes in humans and the great apes. However, the overall MHC class I diversity appears to be low further supporting the hypothesis that gorillas might have experienced a reduction of their MHC repertoire.
Keywords: Evolution; Gogo; Haplotypes; MHC genotyping; Next-generation sequencing; PacBio.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Discovery of gorilla MHC-C expressing C1 ligand for KIR.Immunogenetics. 2018 May;70(5):293-304. doi: 10.1007/s00251-017-1038-y. Epub 2017 Nov 3. Immunogenetics. 2018. PMID: 29101448 Free PMC article.
-
Gorilla class I major histocompatibility complex alleles: comparison to human and chimpanzee class I.J Exp Med. 1991 Dec 1;174(6):1491-509. doi: 10.1084/jem.174.6.1491. J Exp Med. 1991. PMID: 1744581 Free PMC article.
-
Characterization of MHC class II B polymorphism in multiple populations of wild gorillas using non-invasive samples and next-generation sequencing.Am J Primatol. 2015 Nov;77(11):1193-206. doi: 10.1002/ajp.22458. Epub 2015 Aug 17. Am J Primatol. 2015. PMID: 26283172
-
Gorillas with spondyloarthropathies express an MHC class I molecule with only limited sequence similarity to HLA-B27 that binds peptides with arginine at P2.J Immunol. 2001 Mar 1;166(5):3334-44. doi: 10.4049/jimmunol.166.5.3334. J Immunol. 2001. PMID: 11207289 Review.
-
Genetic variation in gorillas.Am J Primatol. 2004 Oct;64(2):161-72. doi: 10.1002/ajp.20070. Am J Primatol. 2004. PMID: 15470746 Review.
Cited by
-
Similar patterns of genetic diversity and linkage disequilibrium in Western chimpanzees (Pan troglodytes verus) and humans indicate highly conserved mechanisms of MHC molecular evolution.BMC Evol Biol. 2020 Sep 15;20(1):119. doi: 10.1186/s12862-020-01669-6. BMC Evol Biol. 2020. PMID: 32933484 Free PMC article.
-
Nomenclature report 2019: major histocompatibility complex genes and alleles of Great and Small Ape and Old and New World monkey species.Immunogenetics. 2020 Feb;72(1-2):25-36. doi: 10.1007/s00251-019-01132-x. Epub 2019 Oct 17. Immunogenetics. 2020. PMID: 31624862
-
Enteroviruses from Humans and Great Apes in the Republic of Congo: Recombination within Enterovirus C Serotypes.Microorganisms. 2020 Nov 13;8(11):1779. doi: 10.3390/microorganisms8111779. Microorganisms. 2020. PMID: 33202777 Free PMC article.
-
MHC class I diversity in chimpanzees and bonobos.Immunogenetics. 2017 Oct;69(10):661-676. doi: 10.1007/s00251-017-0990-x. Epub 2017 Jun 16. Immunogenetics. 2017. PMID: 28623392 Free PMC article.
-
AIDS in chimpanzees: the role of MHC genes.Immunogenetics. 2017 Aug;69(8-9):499-509. doi: 10.1007/s00251-017-1006-6. Epub 2017 Jul 10. Immunogenetics. 2017. PMID: 28695283 Review.
References
-
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, Kimani J, Carrington M, Middleton D, Rajalingam R, Beksac M, Marsh SG, Maiers M, Guethlein LA, Tavoularis S, Little AM, Green RE, Norman PJ, Parham P. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

