The Genetic Code and RNA-Amino Acid Affinities
- PMID: 28333103
- PMCID: PMC5492135
- DOI: 10.3390/life7020013
The Genetic Code and RNA-Amino Acid Affinities
Abstract
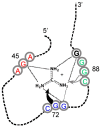
A significant part of the genetic code likely originated via a chemical interaction, which should be experimentally verifiable. One possible verification relates bound amino acids (or perhaps their activated congeners) and ribonucleotide sequences within cognate RNA binding sites. To introduce this interaction, I first summarize how amino acids function as targets for RNA binding. Then the experimental method for selecting relevant RNA binding sites is characterized. The selection method's characteristics are related to the investigation of the RNA binding site model treated at the outset. Finally, real binding sites from selection and also from extant natural RNAs (for example, the Sulfobacillus guanidinium riboswitch) are connected to the genetic code, and by extension, to the evolutionary progression that produced the code. During this process, peptides may have been produced directly on an instructive amino acid binding RNA (a DRT; Direct RNA Template). Combination of observed stereochemical selectivity with adaptation and co-evolutionary refinement is logically required, and also potentially sufficient, to create the striking order conserved throughout the present coding table.
Keywords: DRT; anticodon; binding; codon; triplet.
Conflict of interest statement
The author declares no conflict of interest.
Figures
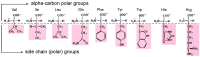

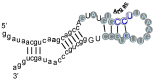
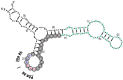
Similar articles
-
RNA-amino acid binding: a stereochemical era for the genetic code.J Mol Evol. 2009 Nov;69(5):406-29. doi: 10.1007/s00239-009-9270-1. Epub 2009 Oct 1. J Mol Evol. 2009. PMID: 19795157 Review.
-
Origins of the genetic code: the escaped triplet theory.Annu Rev Biochem. 2005;74:179-98. doi: 10.1146/annurev.biochem.74.082803.133119. Annu Rev Biochem. 2005. PMID: 15952885 Review.
-
The Uroboros Theory of Life's Origin: 22-Nucleotide Theoretical Minimal RNA Rings Reflect Evolution of Genetic Code and tRNA-rRNA Translation Machineries.Acta Biotheor. 2019 Dec;67(4):273-297. doi: 10.1007/s10441-019-09356-w. Epub 2019 Aug 6. Acta Biotheor. 2019. PMID: 31388859
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
The origin of the genetic code: amino acids as cofactors in an RNA world.Trends Genet. 1999 Jun;15(6):223-9. doi: 10.1016/s0168-9525(99)01730-8. Trends Genet. 1999. PMID: 10354582 Review.
Cited by
-
The Genetic Code Assembles via Division and Fusion, Basic Cellular Events.Life (Basel). 2023 Oct 17;13(10):2069. doi: 10.3390/life13102069. Life (Basel). 2023. PMID: 37895450 Free PMC article.
-
Emergence of a "Cyclosome" in a Primitive Network Capable of Building "Infinite" Proteins.Life (Basel). 2019 Jun 18;9(2):51. doi: 10.3390/life9020051. Life (Basel). 2019. PMID: 31216720 Free PMC article.
-
The Relation Between k-Circularity and Circularity of Codes.Bull Math Biol. 2020 Aug 4;82(8):105. doi: 10.1007/s11538-020-00770-7. Bull Math Biol. 2020. PMID: 32754878 Free PMC article.
-
Chimeric Translation for Mitochondrial Peptides: Regular and Expanded Codons.Comput Struct Biotechnol J. 2019 Aug 23;17:1195-1202. doi: 10.1016/j.csbj.2019.08.006. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31534643 Free PMC article.
-
More Pieces of Ancient than Recent Theoretical Minimal Proto-tRNA-Like RNA Rings in Genes Coding for tRNA Synthetases.J Mol Evol. 2019 Jul;87(4-6):152-174. doi: 10.1007/s00239-019-09892-6. Epub 2019 Apr 5. J Mol Evol. 2019. PMID: 30953098
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

