Environmental and genetic modulation of the phenotypic expression of antibiotic resistance
- PMID: 28333270
- PMCID: PMC5435765
- DOI: 10.1093/femsre/fux004
Environmental and genetic modulation of the phenotypic expression of antibiotic resistance
Abstract
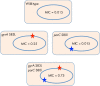
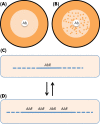
Antibiotic resistance can be acquired by mutation or horizontal transfer of a resistance gene, and generally an acquired mechanism results in a predictable increase in phenotypic resistance. However, recent findings suggest that the environment and/or the genetic context can modify the phenotypic expression of specific resistance genes/mutations. An important implication from these findings is that a given genotype does not always result in the expected phenotype. This dissociation of genotype and phenotype has important consequences for clinical bacteriology and for our ability to predict resistance phenotypes from genetics and DNA sequences. A related problem concerns the degree to which the genes/mutations currently identified in vitro can fully explain the in vivo resistance phenotype, or whether there is a significant additional amount of presently unknown mutations/genes (genetic 'dark matter') that could contribute to resistance in clinical isolates. Finally, a very important question is whether/how we can identify the genetic features that contribute to making a successful pathogen, and predict why some resistant clones are very successful and spread globally? In this review, we describe different environmental and genetic factors that influence phenotypic expression of antibiotic resistance genes/mutations and how this information is needed to understand why particular resistant clones spread worldwide and to what extent we can use DNA sequences to predict evolutionary success.
Keywords: epistasis; heteroresistance; pan genome; persisters; successful clones; virulence.
© FEMS 2017.
Figures

References
-
- Accogli M, Giani T, Monaco M et al. . Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J Antimicrob Chemoth 2014;69:2293–6. - PubMed
-
- Adler M, Anjum M, Andersson DI et al. . Influence of acquired β-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J Antimicrob Chemoth 2012;68:51–9. - PubMed
-
- Adriaenssens N, Coenen S, Versporten A et al. . European Surveillance of Antimicrobial Consumption (ESAC): outpatient quinolone use in Europe (1997-2009). J Antimicrob Chemoth 2011;66:47–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

