Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening
- PMID: 28333914
- PMCID: PMC5526071
- DOI: 10.1038/nprot.2017.016
Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening
Erratum in
-
Author Correction: Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening.Nat Protoc. 2019 Jul;14(7):2259. doi: 10.1038/s41596-018-0063-0. Nat Protoc. 2019. PMID: 30349047
Abstract
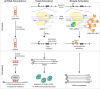
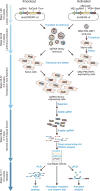
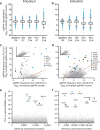
Forward genetic screens are powerful tools for the unbiased discovery and functional characterization of specific genetic elements associated with a phenotype of interest. Recently, the RNA-guided endonuclease Cas9 from the microbial CRISPR (clustered regularly interspaced short palindromic repeats) immune system has been adapted for genome-scale screening by combining Cas9 with pooled guide RNA libraries. Here we describe a protocol for genome-scale knockout and transcriptional activation screening using the CRISPR-Cas9 system. Custom- or ready-made guide RNA libraries are constructed and packaged into lentiviral vectors for delivery into cells for screening. As each screen is unique, we provide guidelines for determining screening parameters and maintaining sufficient coverage. To validate candidate genes identified by the screen, we further describe strategies for confirming the screening phenotype, as well as genetic perturbation, through analysis of indel rate and transcriptional activation. Beginning with library design, a genome-scale screen can be completed in 9-15 weeks, followed by 4-5 weeks of validation.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

