Bioenergetics-based modeling of Plasmodium falciparum metabolism reveals its essential genes, nutritional requirements, and thermodynamic bottlenecks
- PMID: 28333921
- PMCID: PMC5363809
- DOI: 10.1371/journal.pcbi.1005397
Bioenergetics-based modeling of Plasmodium falciparum metabolism reveals its essential genes, nutritional requirements, and thermodynamic bottlenecks
Abstract
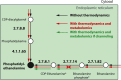
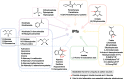
Novel antimalarial therapies are urgently needed for the fight against drug-resistant parasites. The metabolism of malaria parasites in infected cells is an attractive source of drug targets but is rather complex. Computational methods can handle this complexity and allow integrative analyses of cell metabolism. In this study, we present a genome-scale metabolic model (iPfa) of the deadliest malaria parasite, Plasmodium falciparum, and its thermodynamics-based flux analysis (TFA). Using previous absolute concentration data of the intraerythrocytic parasite, we applied TFA to iPfa and predicted up to 63 essential genes and 26 essential pairs of genes. Of the 63 genes, 35 have been experimentally validated and reported in the literature, and 28 have not been experimentally tested and include previously hypothesized or novel predictions of essential metabolic capabilities. Without metabolomics data, four of the genes would have been incorrectly predicted to be non-essential. TFA also indicated that substrate channeling should exist in two metabolic pathways to ensure the thermodynamic feasibility of the flux. Finally, analysis of the metabolic capabilities of P. falciparum led to the identification of both the minimal nutritional requirements and the genes that can become indispensable upon substrate inaccessibility. This model provides novel insight into the metabolic needs and capabilities of the malaria parasite and highlights metabolites and pathways that should be measured and characterized to identify potential thermodynamic bottlenecks and substrate channeling. The hypotheses presented seek to guide experimental studies to facilitate a better understanding of the parasite metabolism and the identification of targets for more efficient intervention.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- World Health Organization; World Malaria Report, 2015. Geneva, Switzerland: WHO, 2016.
-
- Bosi E, Monk JM, Aziz RK, Fondi M, Nizet V, Palsson BO. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci USA. 2016 28 June 2016;113(26):E3801–9. 10.1073/pnas.1523199113 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

