Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions
- PMID: 28334202
- PMCID: PMC6171491
- DOI: 10.1093/bib/bbx008
Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions
Abstract
RNA-Seq is a widely used method for studying the behavior of genes under different biological conditions. An essential step in an RNA-Seq study is normalization, in which raw data are adjusted to account for factors that prevent direct comparison of expression measures. Errors in normalization can have a significant impact on downstream analysis, such as inflated false positives in differential expression analysis. An underemphasized feature of normalization is the assumptions on which the methods rely and how the validity of these assumptions can have a substantial impact on the performance of the methods. In this article, we explain how assumptions provide the link between raw RNA-Seq read counts and meaningful measures of gene expression. We examine normalization methods from the perspective of their assumptions, as an understanding of methodological assumptions is necessary for choosing methods appropriate for the data at hand. Furthermore, we discuss why normalization methods perform poorly when their assumptions are violated and how this causes problems in subsequent analysis. To analyze a biological experiment, researchers must select a normalization method with assumptions that are met and that produces a meaningful measure of expression for the given experiment.
Figures

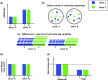
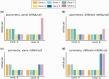

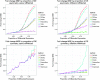
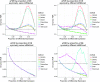
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

