Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots
- PMID: 28334204
- PMCID: PMC5604615
- DOI: 10.1093/aob/mcw256
Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots
Abstract
Background and aims: The root-knot nematode Meloidogyne graminicola is responsible for production losses in rice ( Oryza sativa ) in Asia and Latin America. The accession TOG5681 of African rice, O. glaberrima , presents improved resistance to several biotic and abiotic factors, including nematodes. The aim of this study was to assess the cytological and molecular mechanisms underlying nematode resistance in this accession.
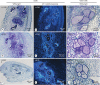
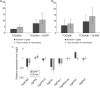
Methods: Penetration and development in M. graminicola in TOG5681 and the susceptible O. sativa genotype 'Nipponbare' were compared by microscopic observation of infected roots and histological analysis of galls. In parallel, host molecular responses to M. graminicola were assessed by root transcriptome profiling at 2, 4 and 8 d post-infection (dpi). Specific treatments with hormone inhibitors were conducted in TOG5681 to assess the impact of the jasmonic acid and salicylic acid pathways on nematode penetration and reproduction.
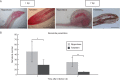
Key results: Penetration and development of M. graminicola juveniles were reduced in the resistant TOG5681 in comparison with the susceptible accession, with degeneration of giant cells observed in the resistant genotype from 15 dpi onwards. Transcriptome changes were observed as early as 2 dpi, with genes predicted to be involved in defence responses, phenylpropanoid and hormone pathways strongly induced in TOG5681, in contrast to 'Nipponbare'. No specific hormonal pathway could be identified as the major determinant of resistance in the rice-nematode incompatible interaction. Candidate genes proposed as involved in resistance to M. graminicola in TOG5681 were identified based on their expression pattern and quantitative trait locus (QTL) position, including chalcone synthase, isoflavone reductase, phenylalanine ammonia lyase, WRKY62 transcription factor, thionin, stripe rust resistance protein, thaumatins and ATPase3.
Conclusions: This study provides a novel set of candidate genes for O. glaberrima resistance to nematodes and highlights the rice- M. graminicola pathosystem as a model to study plant-nematode incompatible interactions.
Keywords: Meloidogyne graminicola; Oryza glaberrima; Oryza sativa; QTL; RNA-Seq; differentially expressed genes; histology; hormones; monocots; resistance responses; root-knot nematode; transcriptomics.
© The Author 2017. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Figures
Similar articles
-
Proteome-Wide Analyses Provide New Insights into the Compatible Interaction of Rice with the Root-Knot Nematode Meloidogyne graminicola.Int J Mol Sci. 2020 Aug 6;21(16):5640. doi: 10.3390/ijms21165640. Int J Mol Sci. 2020. PMID: 32781661 Free PMC article.
-
A Hypersensitivity-Like Response to Meloidogyne graminicola in Rice (Oryza sativa).Phytopathology. 2018 Apr;108(4):521-528. doi: 10.1094/PHYTO-07-17-0235-R. Epub 2018 Mar 5. Phytopathology. 2018. PMID: 29161206
-
QTL mapping for resistance to and tolerance for the rice root-knot nematode, Meloidogyne graminicola.BMC Genet. 2018 Aug 6;19(1):53. doi: 10.1186/s12863-018-0656-1. BMC Genet. 2018. PMID: 30081817 Free PMC article.
-
Meloidogyne graminicola: a major threat to rice agriculture.Mol Plant Pathol. 2017 Jan;18(1):3-15. doi: 10.1111/mpp.12394. Epub 2016 Jul 1. Mol Plant Pathol. 2017. PMID: 26950515 Free PMC article. Review.
-
Root-knot nematodes manipulate plant cell functions during a compatible interaction.J Plant Physiol. 2008 Jan;165(1):104-13. doi: 10.1016/j.jplph.2007.05.007. Epub 2007 Aug 6. J Plant Physiol. 2008. PMID: 17681399 Review.
Cited by
-
Biochemical Defence of Plants against Parasitic Nematodes.Plants (Basel). 2024 Oct 8;13(19):2813. doi: 10.3390/plants13192813. Plants (Basel). 2024. PMID: 39409684 Free PMC article. Review.
-
Combinatorial Interactions of Biotic and Abiotic Stresses in Plants and Their Molecular Mechanisms: Systems Biology Approach.Mol Biotechnol. 2018 Aug;60(8):636-650. doi: 10.1007/s12033-018-0100-9. Mol Biotechnol. 2018. PMID: 29943149 Review.
-
Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops.Plants (Basel). 2025 Apr 27;14(9):1321. doi: 10.3390/plants14091321. Plants (Basel). 2025. PMID: 40364350 Free PMC article. Review.
-
A rice root-knot nematode Meloidogyne graminicola-resistant mutant rice line shows early expression of plant-defence genes.Planta. 2021 Apr 17;253(5):108. doi: 10.1007/s00425-021-03625-0. Planta. 2021. PMID: 33866432
-
Transcriptome analysis of roots from resistant and susceptible rice varieties infected with Hirschmanniella mucronata.FEBS Open Bio. 2019 Nov;9(11):1968-1982. doi: 10.1002/2211-5463.12737. Epub 2019 Oct 22. FEBS Open Bio. 2019. PMID: 31571430 Free PMC article.
References
-
- Agnoun Y, Sié M, Djedatin G. , et al. . 2012. Molecular profiling of interspecific lowland rice progenies resulting from crosses between TOG5681 and TOG5674 (Oryza glaberrima) and IR64 (Oryza sativa). International Journal of Biology 4: 19–28.
-
- Albar L, Ndjiondjop MN, Esshak Z. , et al. . 2003. . Fine genetic mapping of a gene required for rice yellow mottle virus cell-to-cell movement. Theoretical and Applied Genetics 107: 371–378. - PubMed
-
- Albuquerque E, Carneiro R, Costa P. , et al. . 2010. . Resistance to Meloidogyne incognita expresses a hypersensitive-like response in Coffea arabica. European Journal of Plant Pathology 127: 365–373.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

