Basolateral to Central Amygdala Neural Circuits for Appetitive Behaviors
- PMID: 28334609
- PMCID: PMC5480398
- DOI: 10.1016/j.neuron.2017.02.034
Basolateral to Central Amygdala Neural Circuits for Appetitive Behaviors
Abstract
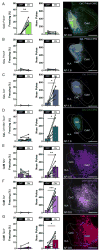
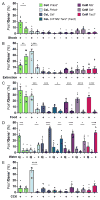
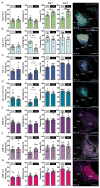
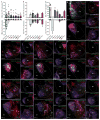
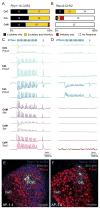
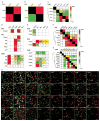
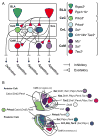
Basolateral amygdala (BLA) principal cells are capable of driving and antagonizing behaviors of opposing valence. BLA neurons project to the central amygdala (CeA), which also participates in negative and positive behaviors. However, the CeA has primarily been studied as the site for negative behaviors, and the causal role for CeA circuits underlying appetitive behaviors is poorly understood. Here, we identify several genetically distinct populations of CeA neurons that mediate appetitive behaviors and dissect the BLA-to-CeA circuit for appetitive behaviors. Protein phosphatase 1 regulatory subunit 1B+ BLA pyramidal neurons to dopamine receptor 1+ CeA neurons define a pathway for promoting appetitive behaviors, while R-spondin 2+ BLA pyramidal neurons to dopamine receptor 2+ CeA neurons define a pathway for suppressing appetitive behaviors. These data reveal genetically defined neural circuits in the amygdala that promote and suppress appetitive behaviors analogous to the direct and indirect pathways of the basal ganglia. VIDEO ABSTRACT.
Keywords: amygdala circuit; appetitive; basolateral amygdala; central amygdala; direct and indirect pathways; drinking; fear; feeding; freezing; reward.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Central amygdala circuits modulate food consumption through a positive-valence mechanism.Nat Neurosci. 2017 Oct;20(10):1384-1394. doi: 10.1038/nn.4623. Epub 2017 Aug 21. Nat Neurosci. 2017. PMID: 28825719
-
Divergent Routing of Positive and Negative Information from the Amygdala during Memory Retrieval.Neuron. 2016 Apr 20;90(2):348-361. doi: 10.1016/j.neuron.2016.03.004. Epub 2016 Mar 31. Neuron. 2016. PMID: 27041499 Free PMC article.
-
Differential efferent projections of GABAergic neurons in the basolateral and central nucleus of amygdala in mice.Neurosci Lett. 2021 Feb 6;745:135621. doi: 10.1016/j.neulet.2020.135621. Epub 2021 Jan 6. Neurosci Lett. 2021. PMID: 33421491
-
New perspectives on central amygdala function.Curr Opin Neurobiol. 2018 Apr;49:141-147. doi: 10.1016/j.conb.2018.02.009. Epub 2018 Mar 6. Curr Opin Neurobiol. 2018. PMID: 29522976 Review.
-
The basolateral amygdala in reward learning and addiction.Neurosci Biobehav Rev. 2015 Oct;57:271-83. doi: 10.1016/j.neubiorev.2015.08.017. Epub 2015 Sep 2. Neurosci Biobehav Rev. 2015. PMID: 26341938 Free PMC article. Review.
Cited by
-
State- and Circuit-Dependent Opponent Processing of Fear.J Neurosci. 2024 Sep 18;44(38):e0857242024. doi: 10.1523/JNEUROSCI.0857-24.2024. J Neurosci. 2024. PMID: 39060174 Free PMC article.
-
Infralimbic medial prefrontal cortex signalling to calbindin 1 positive neurons in posterior basolateral amygdala suppresses anxiety- and depression-like behaviours.Nat Commun. 2022 Sep 17;13(1):5462. doi: 10.1038/s41467-022-33139-6. Nat Commun. 2022. PMID: 36115848 Free PMC article.
-
Sex differences in androgen receptor, estrogen receptor alpha, and c-Fos co-expression with corticotropin releasing factor expressing neurons in restrained adult mice.Horm Behav. 2023 Nov;156:105448. doi: 10.1016/j.yhbeh.2023.105448. Epub 2023 Oct 27. Horm Behav. 2023. PMID: 38344954 Free PMC article.
-
Reconsolidation of a post-ingestive nutrient memory requires mTOR in the central amygdala.Mol Psychiatry. 2021 Jul;26(7):2820-2836. doi: 10.1038/s41380-020-00874-5. Epub 2020 Sep 1. Mol Psychiatry. 2021. PMID: 32873898
-
A dopamine-dependent mechanism for Reward-induced modification of Fear memories.bioRxiv [Preprint]. 2024 Sep 8:2024.09.05.611495. doi: 10.1101/2024.09.05.611495. bioRxiv. 2024. PMID: 39282301 Free PMC article. Preprint.
References
-
- Bandler R, Depaulis A. Elicitation of intraspecific defence reactions in the rat from midbrain periaqueductal grey by microinjection of kainic acid, without neurotoxic effects. Neuroscience letters. 1988;88:291–296. - PubMed
-
- Berridge KC, Robinson TE. Parsing reward. Trends in neurosciences. 2003;26:507–513. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

