N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density
- PMID: 28334758
- PMCID: PMC5449617
- DOI: 10.1093/nar/gkx135
N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density
Abstract
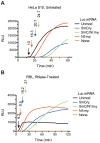
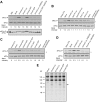
Certain chemical modifications confer increased stability and low immunogenicity to in vitro transcribed mRNAs, thereby facilitating expression of therapeutically important proteins. Here, we demonstrate that N1-methyl-pseudouridine (N1mΨ) outperforms several other nucleoside modifications and their combinations in terms of translation capacity. Through extensive analysis of various modified transcripts in cell-free translation systems, we deconvolute the different components of the effect on protein expression independent of mRNA stability mechanisms. We show that in addition to turning off the immune/eIF2α phosphorylation-dependent inhibition of translation, the incorporated N1mΨ nucleotides dramatically alter the dynamics of the translation process by increasing ribosome pausing and density on the mRNA. Our results indicate that the increased ribosome loading of modified mRNAs renders them more permissive for initiation by favoring either ribosome recycling on the same mRNA or de novo ribosome recruitment.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Sahin U., Kariko K., Tureci O.. mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 2014; 13:759–780. - PubMed
-
- Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011; 29:154–157. - PubMed
-
- Kariko K., Buckstein M., Ni H., Weissman D.. Suppression of Rna recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005; 23:165–175. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

