Amino acid substitution equivalent to human chorea-acanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate
- PMID: 28334785
- PMCID: PMC5393151
- DOI: 10.1093/hmg/ddx054
Amino acid substitution equivalent to human chorea-acanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate
Abstract
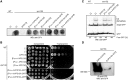
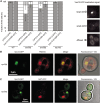
The rare human disorder chorea-acanthocytosis (ChAc) is caused by mutations in hVPS13A gene. The hVps13A protein interacts with actin and regulates the level of phosphatidylinositol 4-phosphate (PI4P) in the membranes of neuronal cells. Yeast Vps13 is involved in vacuolar protein transport and, like hVps13A, participates in PI4P metabolism. Vps13 proteins are conserved in eukaryotes, but their molecular function remains unknown. One of the mutations found in ChAc patients causes amino acids substitution I2771R which affects the localization of hVps13A in skeletal muscles. To dissect the mechanism of pathogenesis of I2771R, we created and analyzed a yeast strain carrying the equivalent mutation. Here we show that in yeast, substitution I2749R causes dysfunction of Vps13 protein in endocytosis and vacuolar transport, although the level of the protein is not affected, suggesting loss of function. We also show that Vps13, like hVps13A, influences actin cytoskeleton organization and binds actin in immunoprecipitation experiments. Vps13-I2749R binds actin, but does not function in the actin cytoskeleton organization. Moreover, we show that Vps13 binds phospholipids, especially phosphatidylinositol 3-phosphate (PI3P), via its SHR_BD and APT1 domains. Substitution I2749R attenuates this ability. Finally, the localization of Vps13-GFP is altered when cellular levels of PI3P are decreased indicating its trafficking within the endosomal membrane system. These results suggest that PI3P regulates the functioning of Vps13, both in protein trafficking and actin cytoskeleton organization. Attenuation of PI3P-binding ability in the mutant hVps13A protein may be one of the reasons for its mislocalization and disrupted function in cells of patients suffering from ChAc.
© The Author 2017. Published by Oxford University Press.
Figures



References
-
- Velayos-Baeza A., Vettori A., Copley R.R., Dobson-Stone C., Monaco A.P. (2004) Analysis of the human VPS13 gene family. Genomics, 84, 536–549. - PubMed
-
- Rampoldi L., Dobson-Stone C., Rubio J.P., Danek A., Chalmers R.M., Wood N.W., Verellen C., Ferrer X., Malandrini A., Fabrizi G.M., et al. (2001) A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet., 28, 119–120. - PubMed
-
- Dobson-Stone C., Velayos-Baeza A., Filippone L.A., Westbury S., Storch A., Erdmann T., Wroe S.J., Leenders K.L., Lang A.E., Dotti M.T., et al. (2004) Chorein detection for the diagnosis of Chorea-acanthocytosis. Ann. Neurol., 56, 299–302. - PubMed
-
- Saiki S., Sakai K., Murata K.Y., Saiki M., Nakanishi M., Kitagawa Y., Kaito M., Gondo Y., Kumamoto T., Matsui M., et al. (2007) Primary skeletal muscle involvement in chorea-acanthocytosis. Mov. Disord., 22, 848–852. - PubMed
-
- Schmidt E.M., Schmid E., Münzer P., Hermann A., Eyrich A.K., Russo A., Walker B., Gu S., Vom Hagen J.M., Faggio C., et al. (2013) Chorein sensitivity of cytoskeletal organization and degranulation of platelets. FASEB J., 27, 2799–2806. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

