H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements
- PMID: 28334823
- PMCID: PMC5449609
- DOI: 10.1093/nar/gkx131
H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements
Abstract
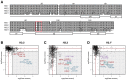
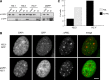
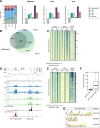
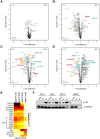
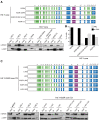
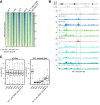
Histone chaperones prevent promiscuous histone interactions before chromatin assembly. They guarantee faithful deposition of canonical histones and functionally specialized histone variants into chromatin in a spatial- and temporally-restricted manner. Here, we identify the binding partners of the primate-specific and H3.3-related histone variant H3.Y using several quantitative mass spectrometry approaches, and biochemical and cell biological assays. We find the HIRA, but not the DAXX/ATRX, complex to recognize H3.Y, explaining its presence in transcriptionally active euchromatic regions. Accordingly, H3.Y nucleosomes are enriched in the transcription-promoting FACT complex and depleted of repressive post-translational histone modifications. H3.Y mutational gain-of-function screens reveal an unexpected combinatorial amino acid sequence requirement for histone H3.3 interaction with DAXX but not HIRA, and for H3.3 recruitment to PML nuclear bodies. We demonstrate the importance and necessity of specific H3.3 core and C-terminal amino acids in discriminating between distinct chaperone complexes. Further, chromatin immunoprecipitation sequencing experiments reveal that in contrast to euchromatic HIRA-dependent deposition sites, human DAXX/ATRX-dependent regions of histone H3 variant incorporation are enriched in heterochromatic H3K9me3 and simple repeat sequences. These data demonstrate that H3.Y's unique amino acids allow a functional distinction between HIRA and DAXX binding and its consequent deposition into open chromatin.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin.Genome Res. 2013 Mar;23(3):440-51. doi: 10.1101/gr.142703.112. Epub 2012 Dec 5. Genome Res. 2013. PMID: 23222847 Free PMC article.
-
The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3.Genes Dev. 2010 Jun 15;24(12):1253-65. doi: 10.1101/gad.566910. Epub 2010 May 26. Genes Dev. 2010. PMID: 20504901 Free PMC article.
-
A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function.J Mol Biol. 2017 Jun 30;429(13):1924-1933. doi: 10.1016/j.jmb.2016.11.010. Epub 2016 Nov 19. J Mol Biol. 2017. PMID: 27871933 Free PMC article. Review.
-
HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes.Cold Spring Harb Symp Quant Biol. 2010;75:27-34. doi: 10.1101/sqb.2010.75.008. Epub 2010 Nov 3. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21047901
-
Daxx and HIRA go viral - How chromatin remodeling complexes affect DNA virus infection.Tumour Virus Res. 2025 Jun;19:200317. doi: 10.1016/j.tvr.2025.200317. Epub 2025 Mar 20. Tumour Virus Res. 2025. PMID: 40120981 Review.
Cited by
-
Chromatin regulation and dynamics in stem cells.Curr Top Dev Biol. 2020;138:1-71. doi: 10.1016/bs.ctdb.2019.11.002. Epub 2019 Dec 30. Curr Top Dev Biol. 2020. PMID: 32220294 Free PMC article. Review.
-
Roles of Histone H2B, H3 and H4 Variants in Cancer Development and Prognosis.Int J Mol Sci. 2024 Sep 7;25(17):9699. doi: 10.3390/ijms25179699. Int J Mol Sci. 2024. PMID: 39273649 Free PMC article. Review.
-
Casting histone variants during mammalian reproduction.Chromosoma. 2023 Sep;132(3):153-165. doi: 10.1007/s00412-023-00803-9. Epub 2023 Jun 22. Chromosoma. 2023. PMID: 37347315 Free PMC article. Review.
-
Epigenetic Regulation of Myogenesis: Focus on the Histone Variants.Int J Mol Sci. 2021 Nov 25;22(23):12727. doi: 10.3390/ijms222312727. Int J Mol Sci. 2021. PMID: 34884532 Free PMC article. Review.
-
Histone variants at a glance.J Cell Sci. 2021 Mar 26;134(6):jcs244749. doi: 10.1242/jcs.244749. J Cell Sci. 2021. PMID: 33771851 Free PMC article. Review.
References
-
- van Holde K.E. Chromatin. 1988; NY: Springer.
-
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. - PubMed
-
- Bonisch C., Nieratschker S.M., Orfanos N.K., Hake S.B.. Chromatin proteomics and epigenetic regulatory circuits. Expert Rev. Proteomics. 2008; 5:105–119. - PubMed
-
- Hake S.B., Garcia B.A., Duncan E.M., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F.. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006; 281:559–568. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

