CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion
- PMID: 28334900
- PMCID: PMC5499812
- DOI: 10.1093/nar/gkx187
CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion
Abstract
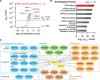
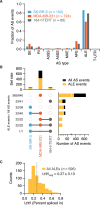
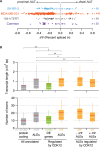
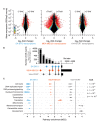
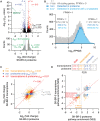
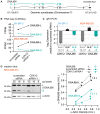
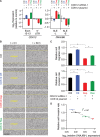
CDK12 (cyclin-dependent kinase 12) is a regulatory kinase with evolutionarily conserved roles in modulating transcription elongation. Recent tumor genome studies of breast and ovarian cancers highlighted recurrent CDK12 mutations, which have been shown to disrupt DNA repair in cell-based assays. In breast cancers, CDK12 is also frequently co-amplified with the HER2 (ERBB2) oncogene. The mechanisms underlying functions of CDK12 in general and in cancer remain poorly defined. Based on global analysis of mRNA transcripts in normal and breast cancer cell lines with and without CDK12 amplification, we demonstrate that CDK12 primarily regulates alternative last exon (ALE) splicing, a specialized subtype of alternative mRNA splicing, that is both gene- and cell type-specific. These are unusual properties for spliceosome regulatory factors, which typically regulate multiple forms of alternative splicing in a global manner. In breast cancer cells, regulation by CDK12 modulates ALE splicing of the DNA damage response activator ATM and a DNAJB6 isoform that influences cell invasion and tumorigenesis in xenografts. We found that there is a direct correlation between CDK12 levels, DNAJB6 isoform levels and the migration capacity and invasiveness of breast tumor cells. This suggests that CDK12 gene amplification can contribute to the pathogenesis of the cancer.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Romano G., Giordano A.. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008; 7:3664–3668. - PubMed
-
- Loyer P., Trembley J.H., Katona R., Kidd V.J., Lahti J.M.. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell Signal. 2005; 17:1033–1051. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

