Mocap: large-scale inference of transcription factor binding sites from chromatin accessibility
- PMID: 28334916
- PMCID: PMC5416775
- DOI: 10.1093/nar/gkx174
Mocap: large-scale inference of transcription factor binding sites from chromatin accessibility
Abstract
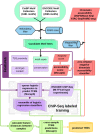
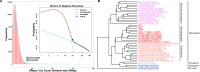
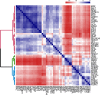
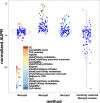
Differential binding of transcription factors (TFs) at cis-regulatory loci drives the differentiation and function of diverse cellular lineages. Understanding the regulatory interactions that underlie cell fate decisions requires characterizing TF binding sites (TFBS) across multiple cell types and conditions. Techniques, e.g. ChIP-Seq can reveal genome-wide patterns of TF binding, but typically requires laborious and costly experiments for each TF-cell-type (TFCT) condition of interest. Chromosomal accessibility assays can connect accessible chromatin in one cell type to many TFs through sequence motif mapping. Such methods, however, rarely take into account that the genomic context preferred by each factor differs from TF to TF, and from cell type to cell type. To address the differences in TF behaviors, we developed Mocap, a method that integrates chromatin accessibility, motif scores, TF footprints, CpG/GC content, evolutionary conservation and other factors in an ensemble of TFCT-specific classifiers. We show that integration of genomic features, such as CpG islands improves TFBS prediction in some TFCT. Further, we describe a method for mapping new TFCT, for which no ChIP-seq data exists, onto our ensemble of classifiers and show that our cross-sample TFBS prediction method outperforms several previously described methods.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Mitchell P.J., Tjian R.. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989; 245:371–378. - PubMed
-
- van Steensel B. Mapping of genetic and epigenetic regulatory networks using microarrays. Nat. Genet. 2005; 37:S18–S24. - PubMed
-
- Junion G., Spivakov M., Girardot C., Braun M., Gustafson E.H., Birney E., Furlong E.E.. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012; 148:473–486. - PubMed
-
- Davidson E. The Regulatory Genome: Gene Regulatory Networks In Development And Evolution. 2010; NY: Elsevier Science.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

