DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation
- PMID: 28334952
- PMCID: PMC5393148
- DOI: 10.1093/hmg/ddx057
DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation
Abstract
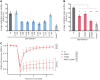
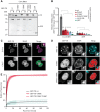
DNMT1 is recruited to substrate sites by PCNA and UHRF1 to maintain DNA methylation after replication. The cell cycle dependent recruitment of DNMT1 is mediated by the PCNA-binding domain (PBD) and the targeting sequence (TS) within the N-terminal regulatory domain. The TS domain was found to be mutated in patients suffering from hereditary sensory and autonomic neuropathies with dementia and hearing loss (HSANIE) and autosomal dominant cerebellar ataxia deafness and narcolepsy (ADCA-DN) and is associated with global hypomethylation and site specific hypermethylation. With functional complementation assays in mouse embryonic stem cells, we showed that DNMT1 mutations P496Y and Y500C identified in HSANIE patients not only impair DNMT1 heterochromatin association, but also UHRF1 interaction resulting in hypomethylation. Similar DNA methylation defects were observed when DNMT1 interacting domains in UHRF1, the UBL and the SRA domain, were deleted. With cell-based assays, we could show that HSANIE associated mutations perturb DNMT1 heterochromatin association and catalytic complex formation at methylation sites and decrease protein stability in late S and G2 phase. To investigate the neuronal phenotype of HSANIE mutations, we performed DNMT1 rescue assays and could show that cells expressing mutated DNMT1 were prone to apoptosis and failed to differentiate into neuronal lineage. Our results provide insights into the molecular basis of DNMT1 dysfunction in HSANIE patients and emphasize the importance of the TS domain in the regulation of DNA methylation in pluripotent and differentiating cells.
© The Author 2017. Published by Oxford University Press.
Figures




References
-
- Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. - PubMed
-
- Goll M.G., Bestor T.H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem., 74, 481–514. - PubMed
-
- Smith Z.D., Meissner A. (2013) DNA methylation: roles in mammalian development. Nat. Rev. Genet., 14, 204–220. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

