Handcuffing reversal is facilitated by proteases and replication initiator monomers
- PMID: 28335002
- PMCID: PMC5397158
- DOI: 10.1093/nar/gkx166
Handcuffing reversal is facilitated by proteases and replication initiator monomers
Abstract
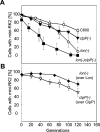
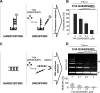
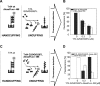
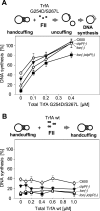
Specific nucleoprotein complexes are formed strictly to prevent over-initiation of DNA replication. An example of those is the so-called handcuff complex, in which two plasmid molecules are coupled together with plasmid-encoded replication initiation protein (Rep). In this work, we elucidate the mechanism of the handcuff complex disruption. In vitro tests, including dissociation progress analysis, demonstrate that the dimeric variants of plasmid RK2 replication initiation protein TrfA are involved in assembling the plasmid handcuff complex which, as we found, reveals high stability. Particular proteases, namely Lon and ClpAP, disrupt the handcuff by degrading TrfA, thus affecting plasmid stability. Moreover, our data demonstrate that TrfA monomers are able to dissociate handcuffed plasmid molecules. Those monomers displace TrfA molecules, which are involved in handcuff formation, and through interaction with the uncoupled plasmid replication origins they re-initiate DNA synthesis. We discuss the relevance of both Rep monomers and host proteases for plasmid maintenance under vegetative and stress conditions.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Ingmer H., Fong E.L., Cohen S.N.. Monomer-dimer equilibrium of the pSC101 RepA protein. J. Mol. Biol. 1995; 250:309–314. - PubMed
-
- Kolatka K., Kubik S., Rajewska M., Konieczny I.. Replication and partitioning of the broad-host-range plasmid RK2. Plasmid. 2010; 64:119–134. - PubMed
-
- Chattoraj D.K. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 2000; 37:467–476. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

