Melting temperature highlights functionally important RNA structure and sequence elements in yeast mRNA coding regions
- PMID: 28335026
- PMCID: PMC5449622
- DOI: 10.1093/nar/gkx161
Melting temperature highlights functionally important RNA structure and sequence elements in yeast mRNA coding regions
Abstract
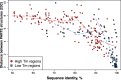
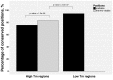
Secondary structure elements in the coding regions of mRNAs play an important role in gene expression and regulation, but distinguishing functional from non-functional structures remains challenging. Here we investigate the dependence of sequence-structure relationships in the coding regions on temperature based on the recent PARTE data by Wan et al. Our main finding is that the regions with high and low thermostability (high Tm and low Tm regions) are under evolutionary pressure to preserve RNA secondary structure and primary sequence, respectively. Sequences of low Tm regions display a higher degree of evolutionary conservation compared to high Tm regions. Low Tm regions are under strong synonymous constraint, while high Tm regions are not. These findings imply that high Tm regions contain thermo-stable functionally important RNA structures, which impose relaxed evolutionary constraint on sequence as long as the base-pairing patterns remain intact. By contrast, low thermostability regions contain single-stranded functionally important conserved RNA sequence elements accessible for binding by other molecules. We also find that theoretically predicted structures of paralogous mRNA pairs become more similar with growing temperature, while experimentally measured structures tend to diverge, which implies that the melting pathways of RNA structures cannot be fully captured by current computational approaches.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Takizawa P.A., DeRisi J.L., Wilhelm J.E., Vale R.D.. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000; 290:341–344. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

