A Community Multi-Omics Approach towards the Assessment of Surface Water Quality in an Urban River System
- PMID: 28335448
- PMCID: PMC5369139
- DOI: 10.3390/ijerph14030303
A Community Multi-Omics Approach towards the Assessment of Surface Water Quality in an Urban River System
Abstract
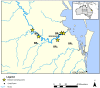
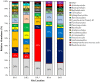
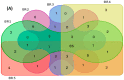
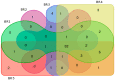
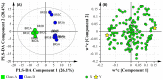
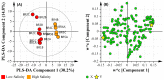
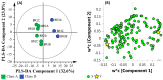
A multi-omics approach was applied to an urban river system (the Brisbane River (BR), Queensland, Australia) in order to investigate surface water quality and characterize the bacterial population with respect to water contaminants. To do this, bacterial metagenomic amplicon-sequencing using Illumina next-generation sequencing (NGS) of the V5-V6 hypervariable regions of the 16S rRNA gene and untargeted community metabolomics using gas chromatography coupled with mass spectrometry (GC-MS) were utilized. The multi-omics data, in combination with fecal indicator bacteria (FIB) counts, trace metal concentrations (by inductively coupled plasma mass spectrometry (ICP-MS)) and in-situ water quality measurements collected from various locations along the BR were then used to assess the health of the river ecosystem. Sites sampled represented the transition from less affected (upstream) to polluted (downstream) environments along the BR. Chemometric analysis of the combined datasets indicated a clear separation between the sampled environments. Burkholderiales and Cyanobacteria were common key factors for differentiation of pristine waters. Increased sugar alcohol and short-chain fatty acid production was observed by Actinomycetales and Rhodospirillaceae that are known to form biofilms in urban polluted and brackish waters. Results from this study indicate that a multi-omics approach enables a deep understanding of the health of an aquatic ecosystem, providing insight into the bacterial diversity present and the metabolic output of the population when exposed to environmental contaminants.
Keywords: chemometrics; contaminated system; metabolomics; metagenomics; trace metals; urban river system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Impact of an urban multi-metal contamination gradient: metal bioaccumulation and tolerance of river biofilms collected in different seasons.Aquat Toxicol. 2015 Feb;159:276-89. doi: 10.1016/j.aquatox.2014.12.014. Epub 2014 Dec 19. Aquat Toxicol. 2015. PMID: 25576823
-
Shift in the microbial community composition of surface water and sediment along an urban river.Sci Total Environ. 2018 Jun 15;627:600-612. doi: 10.1016/j.scitotenv.2018.01.203. Epub 2018 Feb 3. Sci Total Environ. 2018. PMID: 29426184
-
Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies River, South Africa, as a function of land use.Sci Total Environ. 2018 Mar;616-617:326-334. doi: 10.1016/j.scitotenv.2017.10.322. Epub 2017 Nov 7. Sci Total Environ. 2018. PMID: 29126050
-
Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River.J Appl Microbiol. 2013 Nov;115(5):1147-58. doi: 10.1111/jam.12323. Epub 2013 Aug 28. J Appl Microbiol. 2013. PMID: 23924231
-
Toolbox Approaches Using Molecular Markers and 16S rRNA Gene Amplicon Data Sets for Identification of Fecal Pollution in Surface Water.Appl Environ Microbiol. 2015 Oct;81(20):7067-77. doi: 10.1128/AEM.02032-15. Epub 2015 Jul 31. Appl Environ Microbiol. 2015. PMID: 26231650 Free PMC article.
Cited by
-
Omics-based ecosurveillance for the assessment of ecosystem function, health, and resilience.Emerg Top Life Sci. 2022 Apr 15;6(2):185-199. doi: 10.1042/ETLS20210261. Emerg Top Life Sci. 2022. PMID: 35403668 Free PMC article. Review.
-
Current Status of Omics in Biological Quality Elements for Freshwater Biomonitoring.Biology (Basel). 2023 Jun 28;12(7):923. doi: 10.3390/biology12070923. Biology (Basel). 2023. PMID: 37508354 Free PMC article. Review.
-
Microbe-assisted phytoremediation of environmental pollutants and energy recycling in sustainable agriculture.Arch Microbiol. 2021 Dec;203(10):5859-5885. doi: 10.1007/s00203-021-02576-0. Epub 2021 Sep 20. Arch Microbiol. 2021. PMID: 34545411 Review.
-
Detection of Foodborne Pathogens Using Proteomics and Metabolomics-Based Approaches.Front Microbiol. 2018 Dec 18;9:3132. doi: 10.3389/fmicb.2018.03132. eCollection 2018. Front Microbiol. 2018. PMID: 30619201 Free PMC article.
-
Progress and challenges in exploring aquatic microbial communities using non-targeted metabolomics.ISME J. 2023 Dec;17(12):2147-2159. doi: 10.1038/s41396-023-01532-8. Epub 2023 Oct 19. ISME J. 2023. PMID: 37857709 Free PMC article. Review.
References
-
- Robinson A.M., Gondalia S.V., Karpe A.V., Eri R., Beale D.J., Morrison P.D., Palombo E.A., Nurgali K. Fecal microbiota and metabolome in a mouse model of spontaneous chronic colitis: Relevance to human inflammatory bowel disease. Inflamm. Bowel Dis. 2016;22:2767–2787. doi: 10.1097/MIB.0000000000000970. - DOI - PubMed
-
- Kumarasingha R., Karpe A.V., Preston S., Yeo T.C., Lim D.S.L., Tu C.L., Luu J., Simpson K.J., Shaw J.M., Gasser R.B., et al. Metabolic profiling and in vitro assessment of anthelmintic fractions of Picria fel-terrae Lour. Int. J. Parasitol. 2016;6:171–178. doi: 10.1016/j.ijpddr.2016.08.002. - DOI - PMC - PubMed
-
- Bundy J.G., Davey M.P., Viant M.R. Environmental metabolomics: A critical review and future perspectives. Metabolomics. 2009;5:3–21. doi: 10.1007/s11306-008-0152-0. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

