Coccolithoviruses: A Review of Cross-Kingdom Genomic Thievery and Metabolic Thuggery
- PMID: 28335474
- PMCID: PMC5371807
- DOI: 10.3390/v9030052
Coccolithoviruses: A Review of Cross-Kingdom Genomic Thievery and Metabolic Thuggery
Abstract
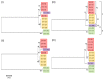
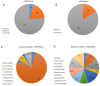
Coccolithoviruses (Phycodnaviridae) infect and lyse the most ubiquitous and successful coccolithophorid in modern oceans, Emiliania huxleyi. So far, the genomes of 13 of these giant lytic viruses (i.e., Emiliania huxleyi viruses-EhVs) have been sequenced, assembled, and annotated. Here, we performed an in-depth comparison of their genomes to try and contextualize the ecological and evolutionary traits of these viruses. The genomes of these EhVs have from 444 to 548 coding sequences (CDSs). Presence/absence analysis of CDSs identified putative genes with particular ecological significance, namely sialidase, phosphate permease, and sphingolipid biosynthesis. The viruses clustered into distinct clades, based on their DNA polymerase gene as well as full genome comparisons. We discuss the use of such clustering and suggest that a gene-by-gene investigation approach may be more useful when the goal is to reveal differences related to functionally important genes. A multi domain "Best BLAST hit" analysis revealed that 84% of the EhV genes have closer similarities to the domain Eukarya. However, 16% of the EhV CDSs were very similar to bacterial genes, contributing to the idea that a significant portion of the gene flow in the planktonic world inter-crosses the domains of life.
Keywords: E. huxleyi; coccolithovirus; domains of life; genome comparison; horizontal gene transfer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wilson W.H., Schroeder D.C., Allen M.J., Holden M.T.G., Parkhill J., Barrell B.G., Churcher C., Hamlin N., Mungall K., Norbertczak H., et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. - DOI - PubMed
-
- Wilson W.H., Tarran G.A., Schroeder D., Cox M.J., Oke J., Malin G. Isolation of viruses responsible for the demise of an Emiliania huxleyi bloom in the English Channel. J. Mar. Biol. Assoc. 2002;82:369–377. doi: 10.1017/S002531540200560X. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

