Opportunistic Sampling of Roadkill as an Entry Point to Accessing Natural Products Assembled by Bacteria Associated with Non-anthropoidal Mammalian Microbiomes
- PMID: 28335605
- PMCID: PMC5368682
- DOI: 10.1021/acs.jnatprod.6b00772
Opportunistic Sampling of Roadkill as an Entry Point to Accessing Natural Products Assembled by Bacteria Associated with Non-anthropoidal Mammalian Microbiomes
Erratum in
-
Correction to Opportunistic Sampling of Roadkill as an Entry Point to Accessing Natural Products Assembled by Bacteria Associated with Non-Anthropoidal Mammalian Microbiomes.J Nat Prod. 2017 Apr 28;80(4):1233. doi: 10.1021/acs.jnatprod.7b00234. Epub 2017 Mar 24. J Nat Prod. 2017. PMID: 28339197 Free PMC article. No abstract available.
Abstract
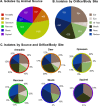
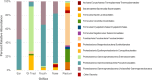
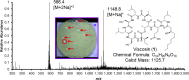
Few secondary metabolites have been reported from mammalian microbiome bacteria despite the large numbers of diverse taxa that inhabit warm-blooded higher vertebrates. As a means to investigate natural products from these microorganisms, an opportunistic sampling protocol was developed, which focused on exploring bacteria isolated from roadkill mammals. This initiative was made possible through the establishment of a newly created discovery pipeline, which couples laser ablation electrospray ionization mass spectrometry (LAESIMS) with bioassay testing, to target biologically active metabolites from microbiome-associated bacteria. To illustrate this process, this report focuses on samples obtained from the ear of a roadkill opossum (Dideiphis virginiana) as the source of two bacterial isolates (Pseudomonas sp. and Serratia sp.) that produced several new and known cyclic lipodepsipeptides (viscosin and serrawettins, respectively). These natural products inhibited biofilm formation by the human pathogenic yeast Candida albicans at concentrations well below those required to inhibit yeast viability. Phylogenetic analysis of 16S rRNA gene sequence libraries revealed the presence of diverse microbial communities associated with different sites throughout the opossum carcass. A putative biosynthetic pathway responsible for the production of the new serrawettin analogues was identified by sequencing the genome of the Serratia sp. isolate. This study provides a functional roadmap to carrying out the systematic investigation of the genomic, microbiological, and chemical parameters related to the production of natural products made by bacteria associated with non-anthropoidal mammalian microbiomes. Discoveries emerging from these studies are anticipated to provide a working framework for efforts aimed at augmenting microbiomes to deliver beneficial natural products to a host.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Analysis of the genomic sequences and metabolites of Serratia surfactantfaciens sp. nov. YD25T that simultaneously produces prodigiosin and serrawettin W2.BMC Genomics. 2016 Nov 3;17(1):865. doi: 10.1186/s12864-016-3171-7. BMC Genomics. 2016. PMID: 27809759 Free PMC article.
-
Correction to Opportunistic Sampling of Roadkill as an Entry Point to Accessing Natural Products Assembled by Bacteria Associated with Non-Anthropoidal Mammalian Microbiomes.J Nat Prod. 2017 Apr 28;80(4):1233. doi: 10.1021/acs.jnatprod.7b00234. Epub 2017 Mar 24. J Nat Prod. 2017. PMID: 28339197 Free PMC article. No abstract available.
-
Broad-spectrum antimicrobial activity of secondary metabolites produced by Serratia marcescens strains.Microbiol Res. 2019 Dec;229:126329. doi: 10.1016/j.micres.2019.126329. Epub 2019 Sep 4. Microbiol Res. 2019. PMID: 31518853
-
Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates.Sci Total Environ. 2022 Sep 10;838(Pt 2):156178. doi: 10.1016/j.scitotenv.2022.156178. Epub 2022 May 23. Sci Total Environ. 2022. PMID: 35618126 Review.
-
Animal Microbiomes as a Source of Novel Antibiotic-Producing Strains.Int J Mol Sci. 2023 Dec 30;25(1):537. doi: 10.3390/ijms25010537. Int J Mol Sci. 2023. PMID: 38203702 Free PMC article. Review.
Cited by
-
Retrospective analysis of natural products provides insights for future discovery trends.Proc Natl Acad Sci U S A. 2017 May 30;114(22):5601-5606. doi: 10.1073/pnas.1614680114. Epub 2017 May 1. Proc Natl Acad Sci U S A. 2017. PMID: 28461474 Free PMC article.
-
Secondary metabolic profiling of Serratia marcescens NP10 reveals new stephensiolides and glucosamine derivatives with bacterial membrane activity.Sci Rep. 2023 Feb 9;13(1):2360. doi: 10.1038/s41598-023-28502-6. Sci Rep. 2023. PMID: 36759548 Free PMC article.
-
Cooperative Involvement of Glycosyltransferases in the Transfer of Amino Sugars during the Biosynthesis of the Macrolactam Sipanmycin by Streptomyces sp. Strain CS149.Appl Environ Microbiol. 2018 Aug 31;84(18):e01462-18. doi: 10.1128/AEM.01462-18. Print 2018 Sep 15. Appl Environ Microbiol. 2018. PMID: 30006405 Free PMC article.
-
From roads to biobanks: Roadkill animals as a valuable source of genetic data.PLoS One. 2023 Dec 7;18(12):e0290836. doi: 10.1371/journal.pone.0290836. eCollection 2023. PLoS One. 2023. PMID: 38060478 Free PMC article.
-
A Metabolomics and Molecular Networking Approach to Elucidate the Structures of Secondary Metabolites Produced by Serratia marcescens Strains.Front Chem. 2021 Mar 16;9:633870. doi: 10.3389/fchem.2021.633870. eCollection 2021. Front Chem. 2021. PMID: 33796505 Free PMC article.
References
-
- Lee M. S.; Oh S.; Tang H. Biomed Mater. Eng. 2014, 24, 3737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

