Widespread promoter methylation of synaptic plasticity genes in long-term potentiation in the adult brain in vivo
- PMID: 28335720
- PMCID: PMC5364592
- DOI: 10.1186/s12864-017-3621-x
Widespread promoter methylation of synaptic plasticity genes in long-term potentiation in the adult brain in vivo
Abstract
Background: DNA methylation is a key modulator of gene expression in mammalian development and cellular differentiation, including neurons. To date, the role of DNA modifications in long-term potentiation (LTP) has not been explored.
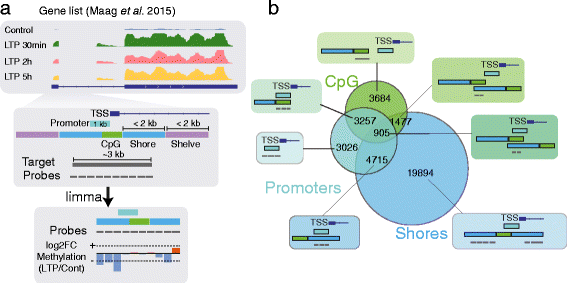
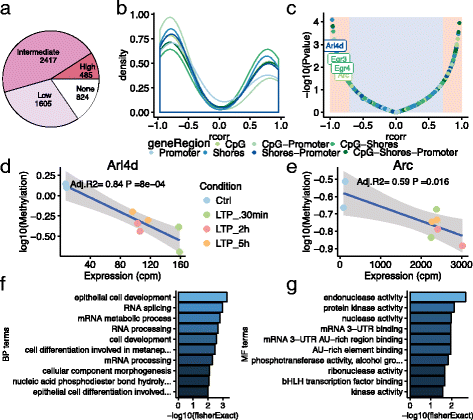
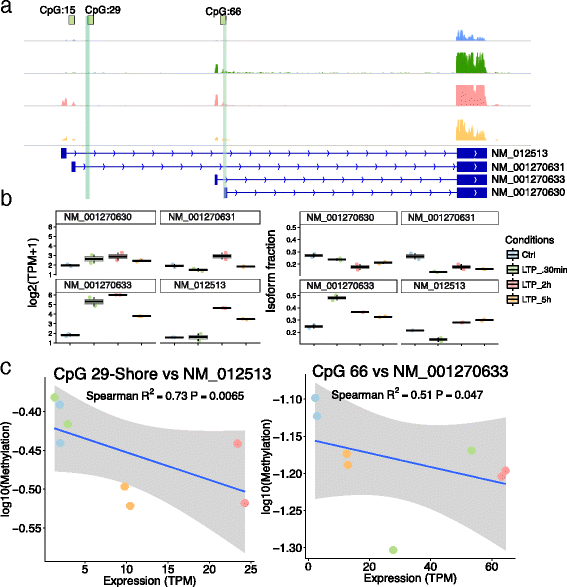
Results: To investigate the occurrence of DNA methylation changes in LTP, we undertook the first detailed study to describe the methylation status of all known LTP-associated genes during LTP induction in the dentate gyrus of live rats. Using a methylated DNA immunoprecipitation (MeDIP)-array, together with previously published matched RNA-seq and public histone modification data, we discover widespread changes in methylation status of LTP-genes. We further show that the expression of many LTP-genes is correlated with their methylation status. We show that these correlated genes are enriched for RNA-processing, active histone marks, and specific transcription factors. These data reveal that the synaptic activity-evoked methylation changes correlates with pre-existing activation of the chromatin landscape. Finally, we show that methylation of Brain-derived neurotrophic factor (Bdnf) CpG-islands correlates with isoform switching from transcripts containing exon IV to exon I.
Conclusions: Together, these data provide the first evidence of widespread regulation of methylation status in LTP-associated genes.
Keywords: DNA-methylation; Epigenetics; LTP; Long-term potentiation; MeDIP; Neuroepigenetics; Synaptic plasticity.
Figures



Similar articles
-
Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex.Neurobiol Learn Mem. 2012 May;97(4):425-40. doi: 10.1016/j.nlm.2012.03.007. Epub 2012 Mar 24. Neurobiol Learn Mem. 2012. PMID: 22469747
-
BDNF-induced local protein synthesis and synaptic plasticity.Neuropharmacology. 2014 Jan;76 Pt C:639-56. doi: 10.1016/j.neuropharm.2013.04.005. Epub 2013 Apr 16. Neuropharmacology. 2014. PMID: 23602987 Review.
-
Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos.Int J Dev Biol. 2010;54(5):803-13. doi: 10.1387/ijdb.103081ll. Int J Dev Biol. 2010. PMID: 20336603
-
Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo.Eur J Neurosci. 2006 Mar;23(6):1501-11. doi: 10.1111/j.1460-9568.2006.04687.x. Eur J Neurosci. 2006. PMID: 16553613
-
BDNF mechanisms in late LTP formation: A synthesis and breakdown.Neuropharmacology. 2014 Jan;76 Pt C:664-76. doi: 10.1016/j.neuropharm.2013.06.024. Epub 2013 Jul 2. Neuropharmacology. 2014. PMID: 23831365 Review.
Cited by
-
Epigenetics in Neurodevelopment: Emerging Role of Circular RNA.Front Cell Neurosci. 2019 Jul 19;13:327. doi: 10.3389/fncel.2019.00327. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31379511 Free PMC article. Review.
-
Epigenome-Wide Association Study of Cognitive Functioning in Middle-Aged Monozygotic Twins.Front Aging Neurosci. 2017 Dec 12;9:413. doi: 10.3389/fnagi.2017.00413. eCollection 2017. Front Aging Neurosci. 2017. PMID: 29311901 Free PMC article.
-
DNA Methylation Dynamics and Cocaine in the Brain: Progress and Prospects.Genes (Basel). 2017 May 12;8(5):138. doi: 10.3390/genes8050138. Genes (Basel). 2017. PMID: 28498318 Free PMC article. Review.
-
Bridging Synaptic and Epigenetic Maintenance Mechanisms of the Engram.Front Mol Neurosci. 2018 Oct 5;11:369. doi: 10.3389/fnmol.2018.00369. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30344478 Free PMC article.
-
A putative role for lncRNAs in epigenetic regulation of memory.Neurochem Int. 2021 Nov;150:105184. doi: 10.1016/j.neuint.2021.105184. Epub 2021 Sep 14. Neurochem Int. 2021. PMID: 34530054 Free PMC article. Review.
References
-
- Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, et al. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci. 2010;31:636–645. doi: 10.1111/j.1460-9568.2010.07112.x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

