Differential gene expression profiling of endometrium during the mid-luteal phase of the estrous cycle between a repeat breeder (RB) and non-RB cows
- PMID: 28335821
- PMCID: PMC5364712
- DOI: 10.1186/s12958-017-0237-6
Differential gene expression profiling of endometrium during the mid-luteal phase of the estrous cycle between a repeat breeder (RB) and non-RB cows
Abstract
Background: Repeat breeding directly affects reproductive efficiency in cattle due to an increase in services per conception and calving interval. This study aimed to investigate whether changes in endometrial gene expression profile are involved in repeat breeding in cows. Differential gene expression profiles of the endometrium were investigated during the mid-luteal phase of the estrous cycle between repeat breeder (RB) and non-RB cows using microarray analysis.
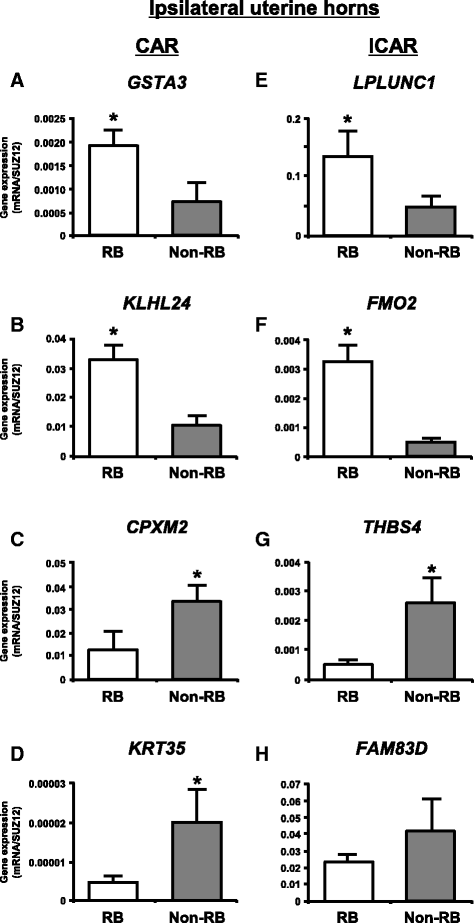
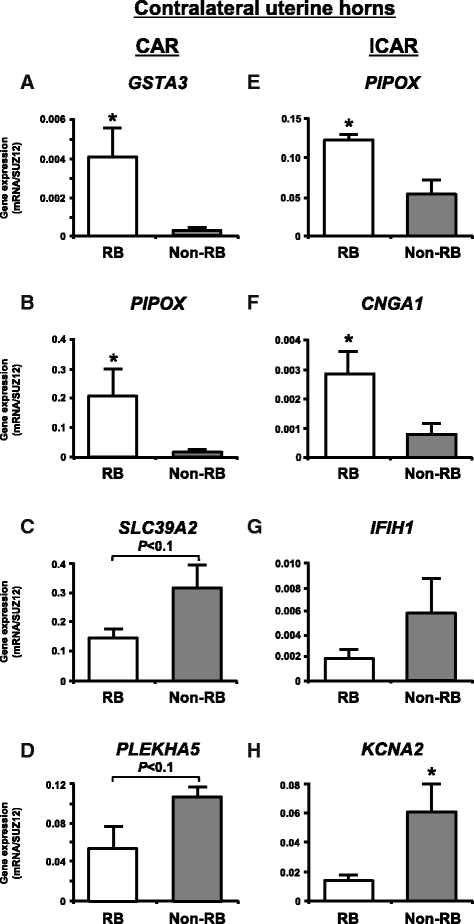
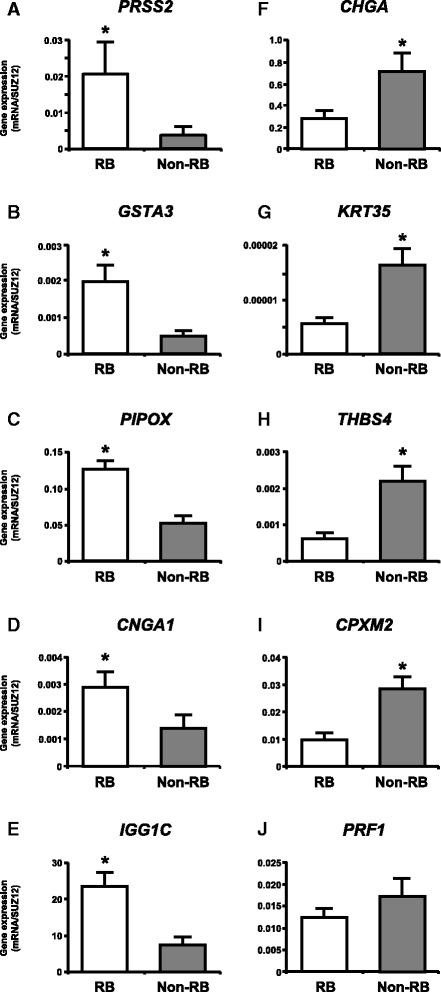
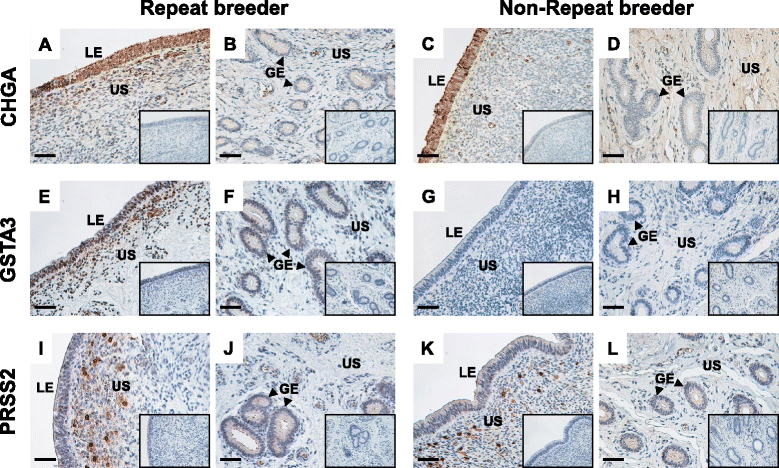
Methods: The caruncular (CAR) and intercaruncular (ICAR) endometrium of both ipsilateral and contralateral uterine horns to the corpus luteum were collected from RB (inseminated at least three times but not pregnant) and non-RB cows on Day 15 of the estrous cycle (4 cows/group). Global gene expression profiles of these endometrial samples were analyzed with a 15 K custom-made oligo-microarray for cattle. Immunohistochemistry was performed to investigate the cellular localization of proteins of three identified transcripts in the endometrium.
Results: Microarray analysis revealed that 405 and 397 genes were differentially expressed in the CAR and ICAR of the ipsilateral uterine horn of RB, respectively when compared with non-RB cows. In the contralateral uterine horn, 443 and 257 differentially expressed genes were identified in the CAR and ICAR of RB, respectively when compared with non-RB cows. Gene ontology analysis revealed that genes involved in development and morphogenesis were mainly up-regulated in the CAR of RB cows. In the ICAR of both the ipsilateral and contralateral uterine horns, genes related to the metabolic process were predominantly enriched in the RB cows when compared with non-RB cows. In the analysis of the whole uterus (combining the data above four endometrial compartments), RB cows showed up-regulation of 37 genes including PRSS2, GSTA3 and PIPOX and down-regulation of 39 genes including CHGA, KRT35 and THBS4 when compared with non-RB cows. Immunohistochemistry revealed that CHGA, GSTA3 and PRSS2 proteins were localized in luminal and glandular epithelial cells and stroma of the endometrium.
Conclusion: The present study showed that endometrial gene expression profiles are different between RB and non-RB cows. The identified candidate endometrial genes and functions in each endometrial compartment may contribute to bovine reproductive performance.
Keywords: Caruncle; Cow; Endometrium; Intercaruncle; Microarray; Repeat breeder.
Figures
Similar articles
-
Differential expression of pro- and anti-angiogenic factors in the endometrium between repeat breeder and normally fertile cows.Anim Reprod Sci. 2023 Jul;254:107265. doi: 10.1016/j.anireprosci.2023.107265. Epub 2023 May 29. Anim Reprod Sci. 2023. PMID: 37270879
-
Exploration of global gene expression changes during the estrous cycle in equine endometrium.Biol Reprod. 2012 Dec 13;87(6):136. doi: 10.1095/biolreprod.112.103226. Print 2012 Jun. Biol Reprod. 2012. PMID: 23077167
-
Expression profiles of perforin, granzyme B and granulysin genes during the estrous cycle and gestation in the bovine endometrium.Anim Sci J. 2014 Jul;85(7):763-9. doi: 10.1111/asj.12209. Epub 2014 May 5. Anim Sci J. 2014. PMID: 24798459
-
Role of chemokines in regulating luteal and uterine functions in pregnant cows.J Reprod Dev. 2024 Jun 1;70(3):145-151. doi: 10.1262/jrd.2023-100. Epub 2024 Feb 28. J Reprod Dev. 2024. PMID: 38403584 Free PMC article. Review.
-
The impact of endometrial scratch performed in mid-luteal phase on the endometrium whole genome transcriptomic profiles in following menstrual cycle.Hum Fertil (Camb). 2023 Oct;26(4):733-741. doi: 10.1080/14647273.2023.2193909. Epub 2023 Apr 8. Hum Fertil (Camb). 2023. PMID: 37029627 Review.
Cited by
-
Inflammatory uterine microenvironment in long-term infertility repeat breeder cows compared with normal fertile cows.Vet Anim Sci. 2024 Jun 1;25:100369. doi: 10.1016/j.vas.2024.100369. eCollection 2024 Sep. Vet Anim Sci. 2024. PMID: 38984268 Free PMC article.
-
Quantitative Proteogenomic Characterization of Inflamed Murine Colon Tissue Using an Integrated Discovery, Verification, and Validation Proteogenomic Workflow.Proteomes. 2022 Apr 14;10(2):11. doi: 10.3390/proteomes10020011. Proteomes. 2022. PMID: 35466239 Free PMC article.
-
Profiles of progesterone and bovine interferon-τ in repeat breeding and non-repeat breeding Aceh cows.Vet World. 2021 Jan;14(1):230-236. doi: 10.14202/vetworld.2021.230-236. Epub 2021 Jan 26. Vet World. 2021. PMID: 33642808 Free PMC article.
-
Using expression data to fine map QTL associated with fertility in dairy cattle.Genet Sel Evol. 2024 Jun 6;56(1):42. doi: 10.1186/s12711-024-00912-8. Genet Sel Evol. 2024. PMID: 38844868 Free PMC article.
-
Seasonal distribution of repeat breeder cows and evaluation of modified protocols for post AI treatment during summer.Trop Anim Health Prod. 2023 Oct 10;55(6):355. doi: 10.1007/s11250-023-03770-6. Trop Anim Health Prod. 2023. PMID: 37816926
References
-
- Perez-Marin CC, Calero GV, Moreno LM. Clinical Approach to the Repeat Breeder Cow Syndrome. In: Perez-Marin CC, editor. A Bird’s-Eye View of Veterinary Medicine. INTECH Open Access Publisher; 2012. doi:10.5772/31374.
-
- Gustafsson H, Larsson K. Embryonic mortality in heifers after artificial insemination and embryo transfer: differences between virgin and repeat breeder heifers. Res Vet Sci. 1985;39:271–274. - PubMed
-
- Albihn A, Gustafsson H, Rodriguez-Martinez H, Larsson K. Development of day 7 bovine demi-embryos transferred into virgin and repeat-breeder heifers. Anim Reprod Sci. 1989;21:161–176. doi: 10.1016/0378-4320(89)90025-0. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

