A conserved NAD+ binding pocket that regulates protein-protein interactions during aging
- PMID: 28336669
- PMCID: PMC5456119
- DOI: 10.1126/science.aad8242
A conserved NAD+ binding pocket that regulates protein-protein interactions during aging
Abstract
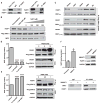
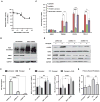
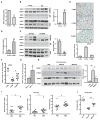
DNA repair is essential for life, yet its efficiency declines with age for reasons that are unclear. Numerous proteins possess Nudix homology domains (NHDs) that have no known function. We show that NHDs are NAD+ (oxidized form of nicotinamide adenine dinucleotide) binding domains that regulate protein-protein interactions. The binding of NAD+ to the NHD domain of DBC1 (deleted in breast cancer 1) prevents it from inhibiting PARP1 [poly(adenosine diphosphate-ribose) polymerase], a critical DNA repair protein. As mice age and NAD+ concentrations decline, DBC1 is increasingly bound to PARP1, causing DNA damage to accumulate, a process rapidly reversed by restoring the abundance of NAD+ Thus, NAD+ directly regulates protein-protein interactions, the modulation of which may protect against cancer, radiation, and aging.
Copyright © 2017, American Association for the Advancement of Science.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

