GmILPA1, Encoding an APC8-like Protein, Controls Leaf Petiole Angle in Soybean
- PMID: 28336772
- PMCID: PMC5462013
- DOI: 10.1104/pp.16.00074
GmILPA1, Encoding an APC8-like Protein, Controls Leaf Petiole Angle in Soybean
Abstract
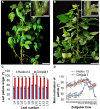
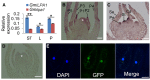
Leaf petiole angle (LPA) is an important plant architectural trait that affects canopy coverage, photosynthetic efficiency, and ultimately productivity in many legume crops. However, the genetic basis underlying this trait remains unclear. Here, we report the identification, isolation, and functional characterization of Glycine max Increased Leaf Petiole Angle1 (GmILPA1), a gene encoding an APC8-like protein, which is a subunit of the anaphase-promoting complex/cyclosome in soybean (Glycine max). A gamma ray-induced deletion of a fragment involving the fourth exon of GmILPA1 and its flanking sequences led to extension of the third exon and formation of, to our knowledge, a novel 3'UTR from intronic and intergenic sequences. Such changes are responsible for enlarged LPAs that are associated with reduced motor cell proliferation in the Gmilpa1 mutant. GmILPA1 is mainly expressed in the basal cells of leaf primordia and appears to function by promoting cell growth and division of the pulvinus that is critical for its establishment. GmILPA1 directly interacts with GmAPC13a as part of the putative anaphase-promoting complex. GmILPA1 exhibits variable expression levels among varieties with different degrees of LPAs, and expression levels are correlated with the degrees of the LPAs. Together, these observations revealed a genetic mechanism modulating the plant petiole angle that could pave the way for modifying soybean plant architecture with optimized petiole angles for enhanced yield potential.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Cheng W, Gao JS, Feng XX, Shao Q, Yang SX, Feng XZ (2016) Characterization of dwarf mutants and molecular mapping of a dwarf locus in soybean. J Integr Agric 15: 60345–60347
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

